Enzyme Substrates
Products for Enzyme Substrates
- Cat.No. Product Name Information
-
GC42151
2'-Deoxyguanosine 5'-monophosphate (sodium salt hydrate)
dGMP
2'-Deoxyguanosine 5'-monophosphate (dGMP) is used as a substrate of guanylate kinases to generate dGDP, which in turn is phosphorylated to dGTP, a nucleotide precursor used in DNA synthesis.
-
GC49272
2-Chloro-4-nitrophenyl α-D-maltotrioside
CNP-G3
A colorimetric α-amylase substrate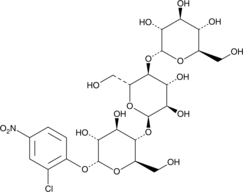
-
GC42137
2-Chloro-4-nitrophenyl-α-D-glucopyranoside
CNPαDglucopyranoside
Carbohydrates conjugated with 2-chloro-4-nitrophenyl (CNP) serve as chromogenic substrates in assays for enzymes that release CNP from the conjugated carbohydrate.
-
GC41637
3'-Deoxyguanosine
3'-Deoxyguanosine is a ligand that can be complexed with enzymes, such as purine nucleoside phosphorylase, and receptors, in order to study structure-activity relationships.

-
GC42290
3-Hydroxyglutaric Acid dimethyl ester
Dimethyl 3-HG, Dimethyl β-HG, Dimethyl 3-hydroxyglutarate, Dimethyl 3-hydroxypentanedioate, 3-HG dimethyl ester, β-HG dimethyl ester, NSC 30047
3-Hydroxyglutaric acid dimethyl ester is a prochiral substrate and an esterified derivative of 3-hydroxyglutaric acid.
-
GC49121
3-Indoxyl Phosphate (sodium salt)
3-IP
A chromogenic and electrochemical substrate for alkaline phosphatase
-
GC48983
4-Aminophenyl Phosphate (sodium salt hydrate)
An alkaline phosphatase substrate

-
GC49114
4-Methylumbelliferyl-β-D-Galactoside
4-Methylumbelliferyl-β-D-Galactopyranoside
4-Methylumbelliferyl-β-D-Galactoside is a fluorescent substrate for β-galactosidase which, when cleaved, produces a water-soluble blue fluorescent coumarin fluorophore that can be detected using a fluoroenzymeter or fluorometer.
-
GC45913
4-Nitrophenyl α-L-rhamnopyranoside
p-Nitrophenyl α-L-rhamnoside

-
GC42461
4-Nitrophenyl β-D-Cellobioside
p-Nitrophenyl β-D-Cellobioside, para-Nitrophenyl β-D-Cellobioside
4-Nitrophenyl β-D-cellobioside is a disaccharide and an enzyme substrate.
-
GC42462
4-Nitrophenyl β-D-Cellotrioside
p-Nitrophenyl β-D-Cellotrioside
4-Nitrophenyl β-D-cellotrioside is a chromogenic substrate for endoglucanases and cellobiohydrolases.
-
GC40089
4-Nitrophenyl Butyrate
p-Nitrophenyl Butyrate, para-Nitrophenyl Butyrate, 4-NPB, NSC 6867
4-Nitrophenyl butyrate (4-NPB) is a colorimetric substrate for lipases.
-
GC46672
4-Nitrophenyl Palmitate
p-Nitrophenyl Palmitate, para-Nitrophenyl Palmitate, pNpp
A colorimetric lipase and esterase substrate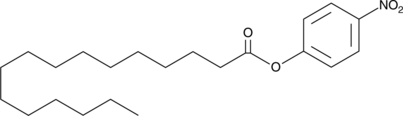
-
GC49158
4-Nitrophenyl Phosphate (sodium salt hydrate)
p-Nitrophenyl Phosphate, para-Nitrophenyl Phosphate, 4-NPP, pNPP
4-Nitrophenyl phosphate (p-nitrophenyl phosphate) disodium hexahydrate is widely used as a small molecule phosphotyrosine-like substrate in activity assays for protein tyrosine phosphatases.
-
GC18209
4-Nitrophenyl β-D-Cellotetraoside
p-Nitrophenyl β-D-Cellotetraoside
4-Nitrophenyl β-D-cellotetraoside is small molecule cellulose mimic that consists of a tetramer of D-glucose units joined by β-1-4-glycosidic bonds.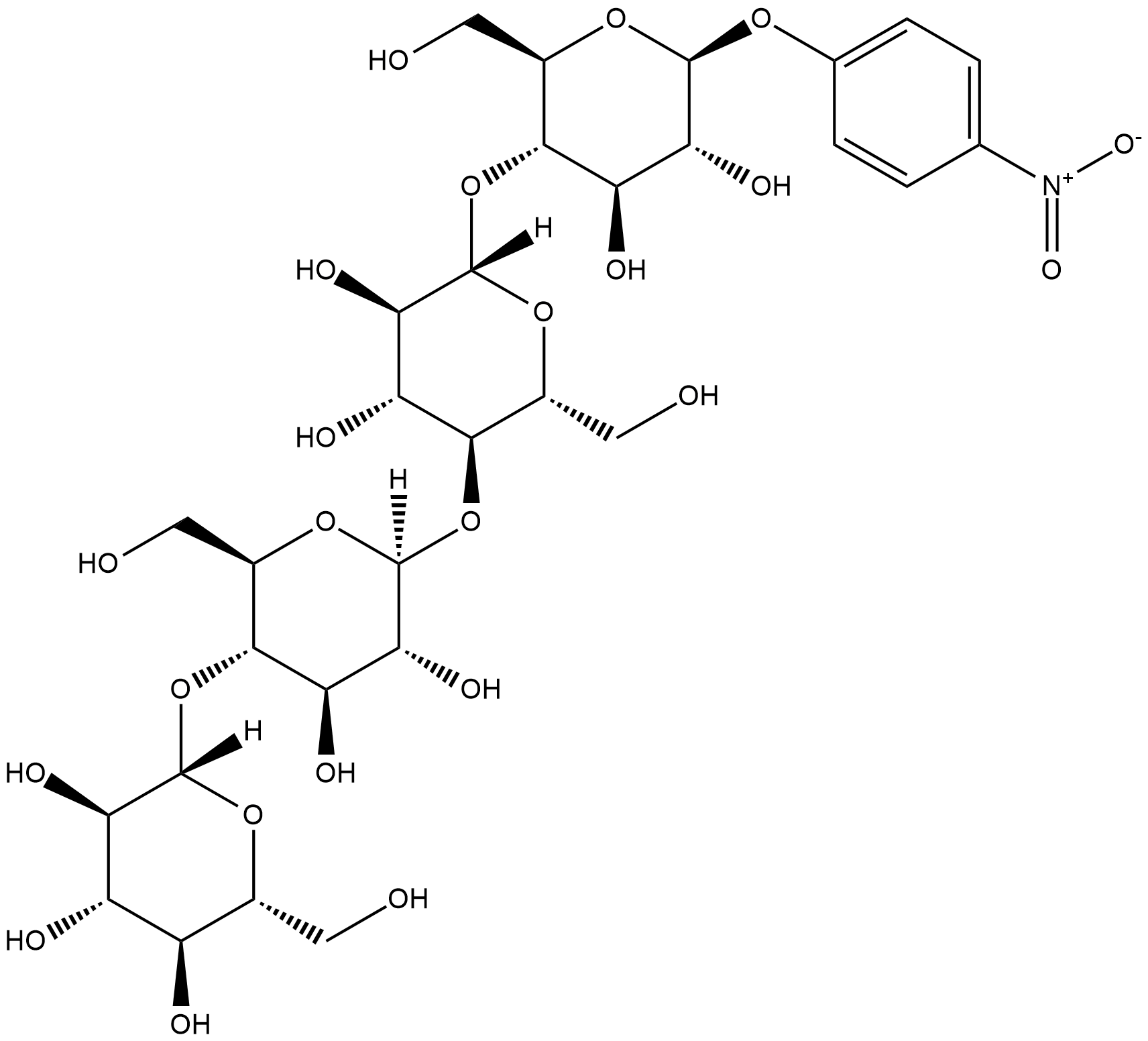
-
GC42463
4-Nitrophenyl-N-acetyl-α-D-galactosaminide
pNP-GlcNAc
N-acetyl-D-galactosaminidase catalyzes the cleavage of N-acetylglucosamine (GlcNAc), the monomeric unit of the polymer chitin.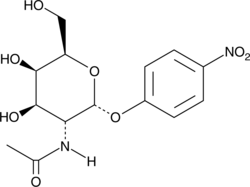
-
GC45841
4-Nitrophenyl-N-acetyl-β-D-glucosaminide
GlcNAc-PNP, p-Nitrophenyl-N-acetyl-β-D-glucosaminide
A chromogenic substrate for N-acetyl-β-glucosaminidase
-
GC46722
6-Chloro-3-indolyl-β-D-Glucuronide (cyclohexylammonium salt)
A β-glucuronidase chromogenic substrate

-
GC49320
7-Ethoxy-4-(trifluoromethyl)coumarin
EFC
A fluorogenic substrate for CYPs
-
GC49030
7-Ethoxycoumarin
A CYP450 substrate

-
GC49248
7-Methoxy-4-(trifluoromethyl)coumarin
MFC
A fluorogenic substrate for CYPs
-
GC91462
Abl Substrate Peptide (trifluoroacetate salt)
Abl substrate peptide is a peptide substrate for the tyrosine kinase Abl.

-
GA20494
Ac-Asp-Glu-Val-Asp-pNA
Ac-Asp-Glu-Val-Asp-pNA
The cleavage of the chromogenic caspase-3 substrate Ac-DEVD-pNA can be monitored at 405 nm.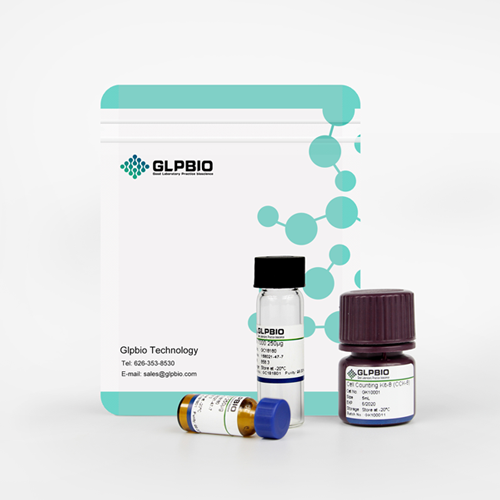
-
GA20507
Ac-D-Leu-OH
Ac-D-Leu-OH is an amino protecting group N-substituted chiral amino acid.

-
GC49725
Ac-IETD-AFC (trifluoroacetate salt)
N-Acetyl-Ile-Glu-Thr-Asp-7-amino-4-Trifluoromethylcoumarin
A fluorogenic substrate for caspase-8
-
GC48974
Ac-VEID-AMC (ammonium acetate salt)
NAcetylValGluIleAsp7amido4Methylcoumarin, Caspase6 Substrate (Fluorogenic)
A caspase-6 fluorogenic substrate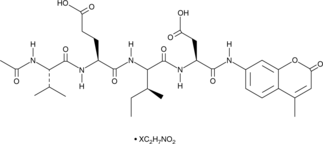
-
GC49717
Boc-LGR-pNA (acetate)
Boc-Leu-Gly-Arg-pNA, Boc-Leu-Gly-Arg-p-Nitroanilide, Tertbutyloxycarbonyl-Leu-Gly-Arg-p-Nitroanilide
A chromogenic substrate for endotoxins
-
GC45394
Bz-IEGR-pNA (acetate)
Bz-Ile-Glu-Gly-Arg-pNA, Bz-Ile-Glu-Gly-Arg-p-nitroanilide, Factor Xa Chromogenic Substrate

-
GC49332
Calcineurin Substrate (trifluoroacetate salt)
H-Asp-Leu-Asp-Val-Pro-Ile-Pro-Gly-Arg-Phe-Asp-Arg-Arg-Val-Ser-Val-Ala-Ala-Glu-OH, H-DLDVPIPGRFDRRVSVAAE-OH
A synthetic peptide substrate
-
GC47080
Chlorophenol Red β-D-Galactopyranoside
CPRG
Chlorophenol Red β-D-Galactopyranoside is a long-wavelength dye, which has been widely used for colorimetric assays.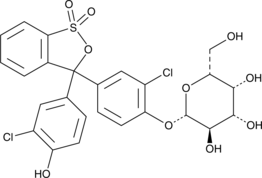
-
GC43392
Decanoyl m-Nitroaniline
DemNA
Decanoyl m-Nitroaniline (DemNA) is one of several nitroaniline fatty acid amides which can be used to measure fatty acid amide hydrolase (FAAH) activity.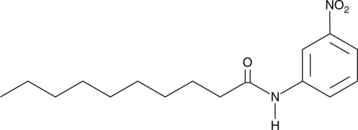
-
GC48995
Enteropeptidase Fluorogenic Substrate (trifluoroacetate salt)
A fluorogenic substrate of enteropeptidase

-
GC91428
FALGPA (trifluoroacetate salt)
2-Furanacryloyl-L-leucylglycyl-L-prolyl-L-alanine; 2-Furanacryloyl-Leu-Gly-Pro-Ala-OH
FALGPA is a colorimetric substrate for collagenase.
-
GC18201
Gly-Pro-pNA (hydrochloride)
Gly-Pro pnitroanilide, GP-pNA
Gly-Pro-pNA (hydrochloride) is a chromogenic substrate that can be cleaved by the circulating enzyme, dipeptidyl peptidase IV (DPP IV).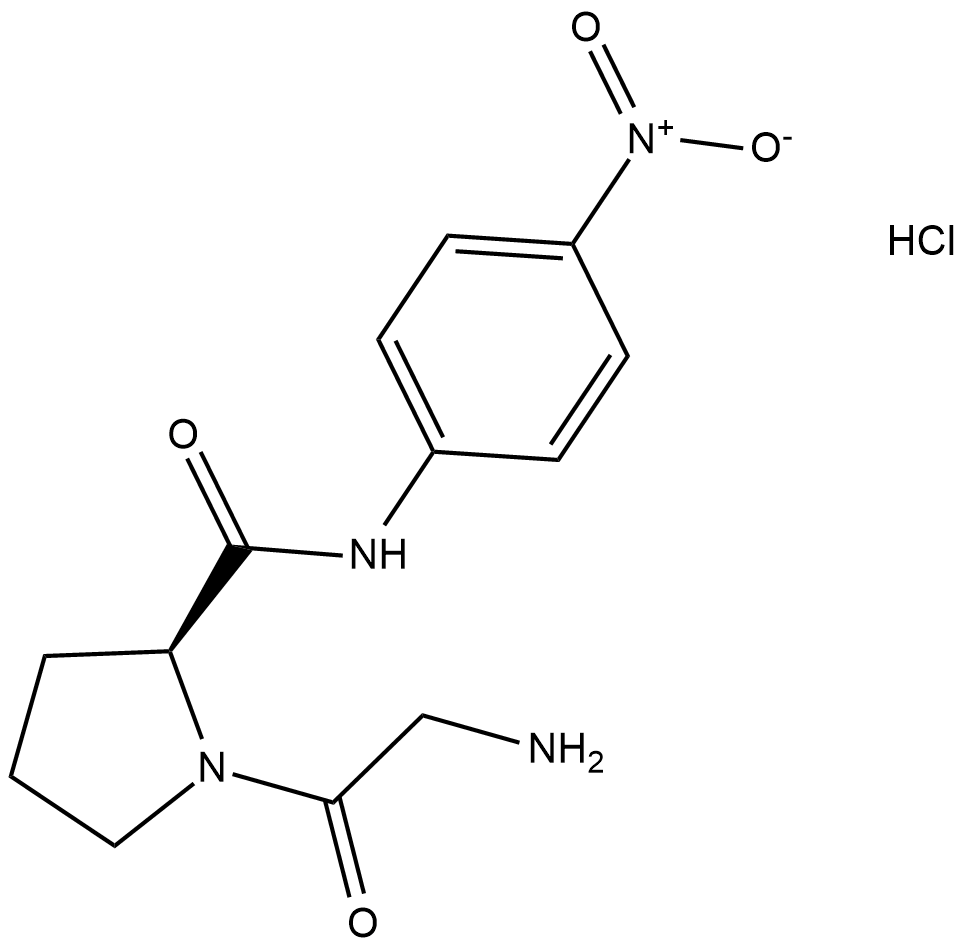
-
GA22303
H-D-Pro-Phe-Arg-pNA . 2 HCl
pFR-pNA, Plasma Kallikrein Chromogenic Substrate, D-Pro-Phe-Arg-p-nitroaniline
pFR-pNA, chromogenic substrate for the determination of plasma kallikrein-like activity. CAS Number (net): 64816-19-9.
-
GC45471
H-Gly-Arg-pNA (hydrochloride)
Gly-Arg-4-NA, GR-pNA, GR p-nitroanilide

-
GC43914
Isobutyryl Coenzyme A (sodium salt)
Isobutyryl-CoA
Isobutyryl coenzyme A (isobutyryl-CoA) is a short-chain branched acyl CoA.
-
GC49547
L-Glutamic Acid γ-p-Nitroanilide (hydrate)
γ-Glu-pNA, Glutamate γ-(4-Nitroanilide), L-GPNA, E-pNA
A colorimetric substrate for γ-glutamyl transpeptidase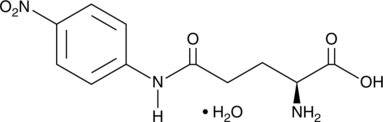
-
GC49205
L-Leu-AMC (hydrochloride)
Leu-MCA, L-Leucine-7-amido-4-methylcoumarin, 7-L-Leucyl-4-methylcoumarinylamide
L-Leucine-7-amido-4-methylcoumarin (Leu-AMC) hydrochloride is a bright blue fluorogenic peptidyl substrate for LAP3 (leucine aminopeptidase).
-
GC44156
MeOSuc-AAPV-pNA
MeOSuc-AAPV-pNA is a sensitive fluorogenic substrate of human leukocyte and porcine pancreatic elastase.

-
GC45667
N-(p-Tosyl)-GPR-pNA (acetate)
N-(p-Tosyl)-Gly-Pro-Arg-p-Nitroanilide, Tos-Gly-Pro-Arg-pNA
A colorimetric thrombin substrate
-
GC44396
N-Hippuryl-His-Leu (hydrate)
N-Benzoyl-Gly-His-Leu
N-Hippuryl-His-Leu (hydrate) is a synthetic substrate for angiotensin-converting enzyme (ACE) that has been used in the in vitro identification of ACE inhibitors.,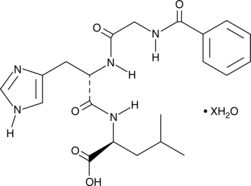
-
GC47806
N-p-Tosyl-Gly-Pro-Lys-pNA (acetate)
N-p-Tosyl-Gly-Pro-Lys-p-nitroanilide
A colorimetric plasmin substrate
-
GC44285
N1-Acetylspermine (hydrochloride)
N1-Acetylspermine is a monoacetylated derivative of spermine, an endogenous polyamine synthesized from spermidine, that displays lower Km and higher Vmax values than spermine, making it a better substrate of polyamine oxidase than the non-acetylated polyamine.

-
GC47739
NADH (sodium salt hydrate)
Nicotinamide adenine dinucleotide, reduced
A reduced form of NAD+
-
GC49501
Nirvanol
rac-N-Desmethyl Mephenytoin, NSC 150466, NSC 33388, Phenylethylhydantoin
An active metabolite of mephenytoin
-
GC14823
Nitrocefin
Nitrocephin
Used for detection of β-lactamases

-
GC44511
O-Phospho-L-Tyrosine
Phosphotyrosine, P-Tyr
O-Phospho-L-tyrosine is a phosphorylated form of L-tyrosine.
-
GC44664
p-Nitrophenylphosphorylcholine
4-Nitrophenylphosphorylcholine, O-(4-Nitrophenylphosphoryl)choline, para-Nitrophenylphosphorylcholine, 4-NPPC, p-NPPC, para-NPPC
p-Nitrophenylphosphorylcholine is a chromogenic substrate that is used to measure phospholipase C (PLC) activity.
-
GC44672
PPHP
PPHP is a substrate for the measurement of peroxidase enzymes.

-
GC44907
S-NEPC
Cytochrome P450 metabolites of arachidonic acid, such as 11(12)-EpETrE and 14(15)-EpETrE have been identified as endothelium derived hyperpolarizing factors with vasodilator activity.

-
GC18565
Suc-AAP-Abu-pNA
Elastase Colorimetric Substrate IV
Suc-AAP-Abu-pNA is a chromogenic peptide substrate for pancreatic elastase.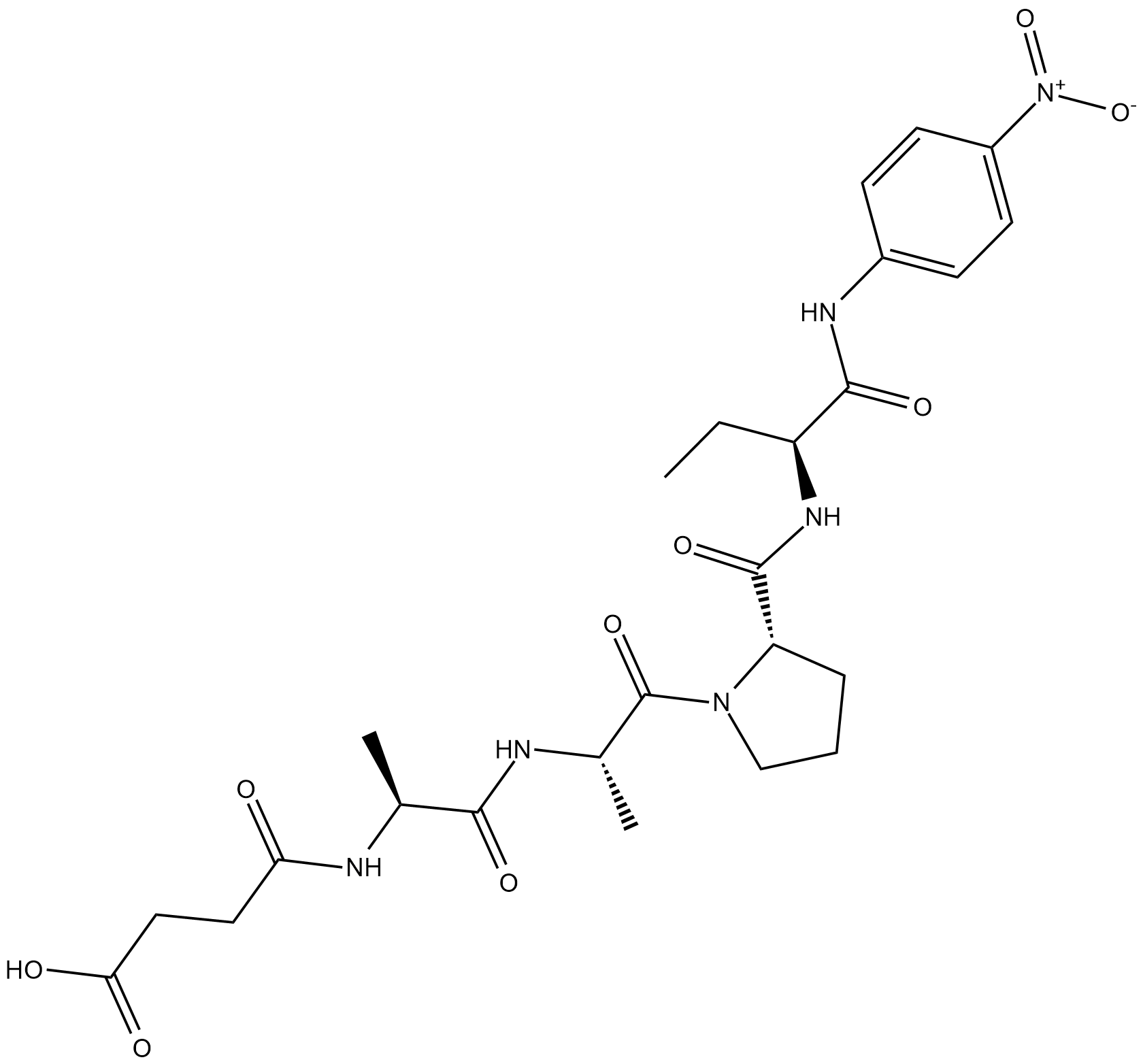
-
GC91437
Suc-AAP-Abu-pNA (trifluoroacetate salt)
Elastase Colorimetric Substrate IV
Suc-AAP-Abu-pNA is a chromogenic peptide substrate for pancreatic elastase.
-
GC44960
Suc-AAPF-pNA
SuccinylAlaAlaProPhepnitroanilide
Suc-AAPF-pNA (Suc-Ala-Ala-Pro-Phe-pNA) is a colorimetrically specific substrate for elastase. Under the action of enzymes, Suc-AAPF-pNA is hydrolyzed, releasing yellow p-nitroaniline (pNA), and the change in absorbance at 400-410 nm is measured to reflect enzyme activity through colorimetry.
-
GA23570
Suc-Tyr-Val-Ala-Asp-pNA
Caspase-1 and Caspase-4 Chromogenic Substrate, Suc-Tyr-Val-Ala-Asp-p-nitroanilide
Suc-YVAD-pNA, a chromogenic caspase-1 substrate.
-
GA23742
Z-D-Arg-Gly-Arg-pNA . 2 HCl
Benzyl-D-Arg-Gly-Arg-pNA, Benzyl-D-Arg-Gly-Arg-p-nitroanilide, Factor Xa Chromogenic Substrate, S-2765
Z-RGR-pNA is a chromogenic substrate for the determination of factor Xa.
-
GC49145
Z-FR-AMC (trifluoroacetate salt)
Z-FR-7-amino-4-Methylcoumarin, Z-Phe-Arg-AMC
A fluorogenic substrate for cathepsin L


