- Cat.No. Product Name Information
-
GC26360
SYTO 9 (20x in DMSO) Nucleic Acid Dyes for PCR
SYTO9 Gren fluorescent Nucleic Acid Stain (SVTO9 Gren fluorescent Nucleic Acid Stain) is an excellent counterstain for nuclei and chromosomes and is highly permeable to cell membranes.

-
GC26359
Biotin-labeled Agatolimod sodium
Biotin-labeled Agatolimod (sodium), a class B CpG ODN (oligodeoxynucleotide), is a TLR9 agonist.

-
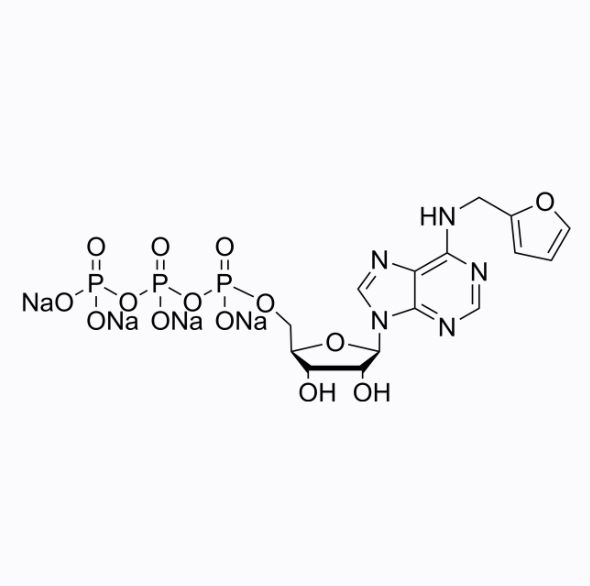
GC26358
Kinetin triphosphate tetrasodium
Kinetin triphosphate (6-Fu ATP) tetrasodium is an ATP analogue used to regulate or enhance kinase function, and its catalytic efficiency is higher than its endogenous substrate ATP. Kinetin triphosphate tetrasorium can be used for research on Parkinson's disease.
-
GC26357
ODN M362
ODN M362 is a C-class oligonucleotide that is a TLR-9 agonist and can be used as a vaccine adjuvant. ODN M362 induces apoptosis in cancer cells.

-
GC26356
Sodium fluoride, 99.99%metals basis

-
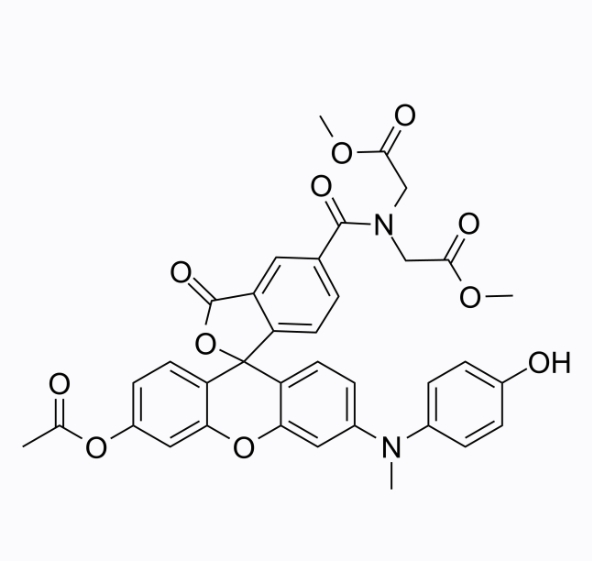
GC26355
HKGreen-4I
HKGreen-4I is a green fluorescent probe that selectively binds ONOO- in living cells with a maximum excitation/emission wavelength of 520/543nm.
-
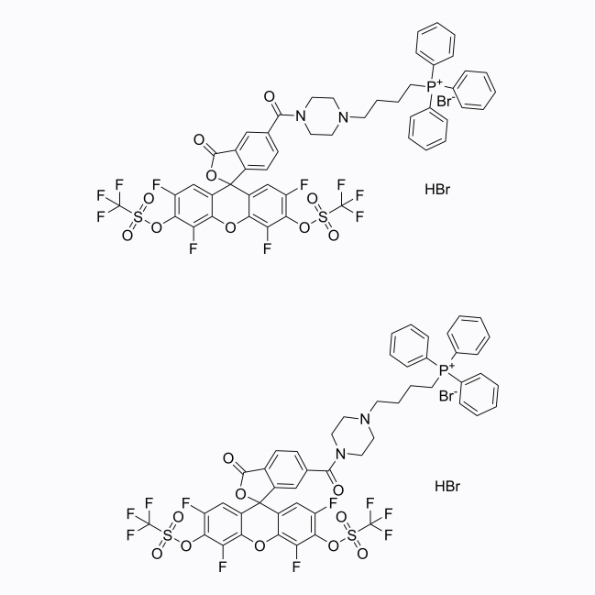
GC26354
HKSOX-1m (5/6-mixture) (hydrobromide)
HKSOX-1m (5/6-mixture) (hydrobromide) is a reactive oxygen species fluorescent probe targeting mitochondria, with a maximum absorption/emission wavelength of 509/534nm.
-
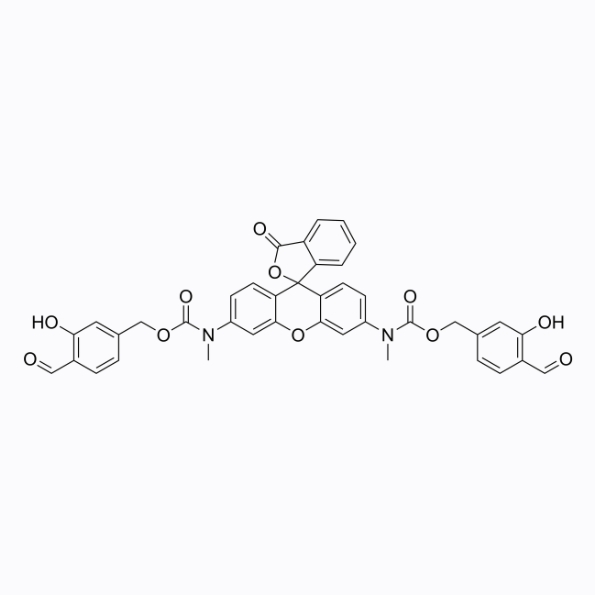
GC26353
HKPerox-1
HKPerox-1 is a live cell green fluorescent probe that specifically binds to intracellular hydroxyl radicals(·OH) with a maximum absorption/emission wavelength of 520/543nm.
-
GC26352
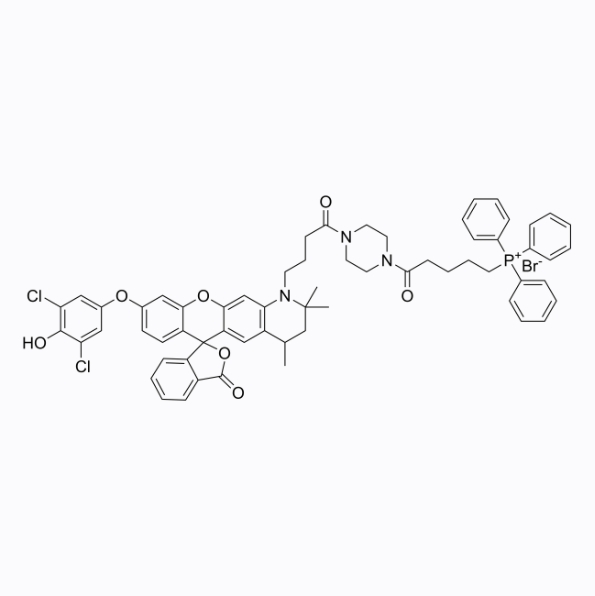
HKOCl-4m
HKOCl-4m is a living cell hypochlorous acid fluorescent probe with a maximum absorption/emission wavelength of 490/527nm.
-
GC26351
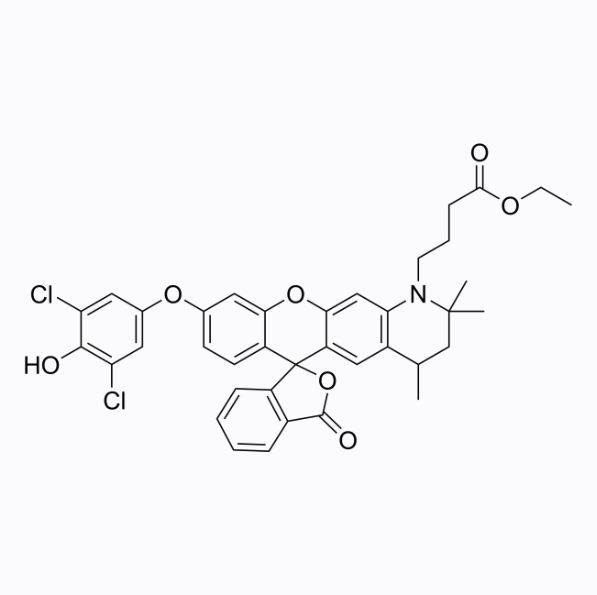
HKOCl-4
HKOCl-4 is a living cell hypochlorous acid fluorescent probe with a maximum absorption/emission wavelength of 530/557nm.
-
GC26350
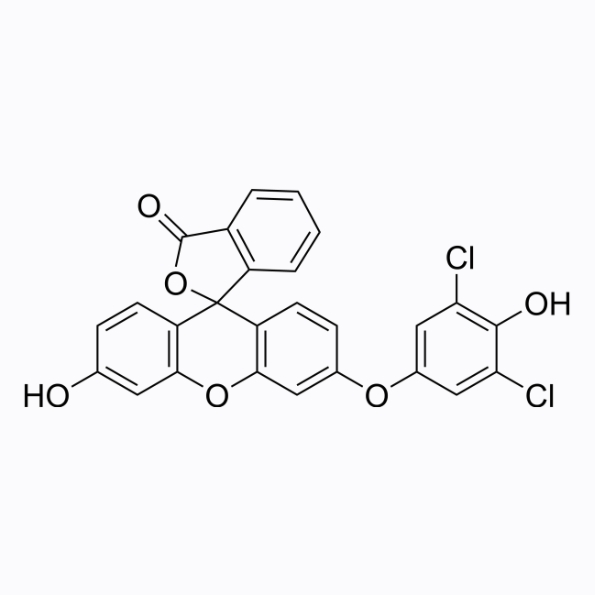
HKOCl-3
HKOCl-3 is a living cell hypochlorous acid fluorescent probe with a maximum absorption/emission wavelength of 490/527nm.
-
GC26349
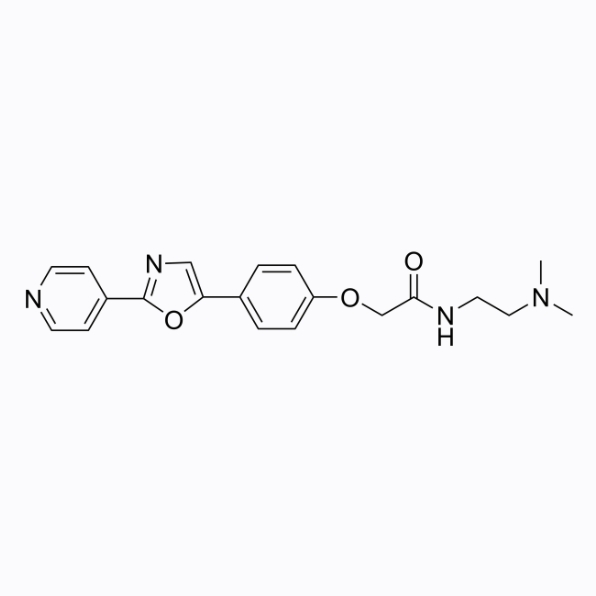
LysoSensor PDMPO
PDMPO is an acidophilic lysosomal fluorescent probe and intracellular pH indicator that can be used for fluorescence imaging
-
GC26348
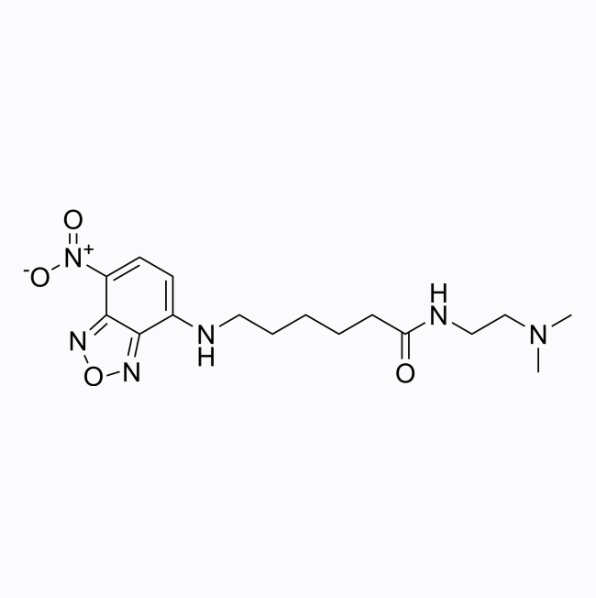
LysoTracker Yellow HCK 123
LysoTracker Yellow HCK 123 is a cell-penetrating lysosomal fluorescent probe with a maximum excitation/emission light of 465/535nm.
-
GC26347
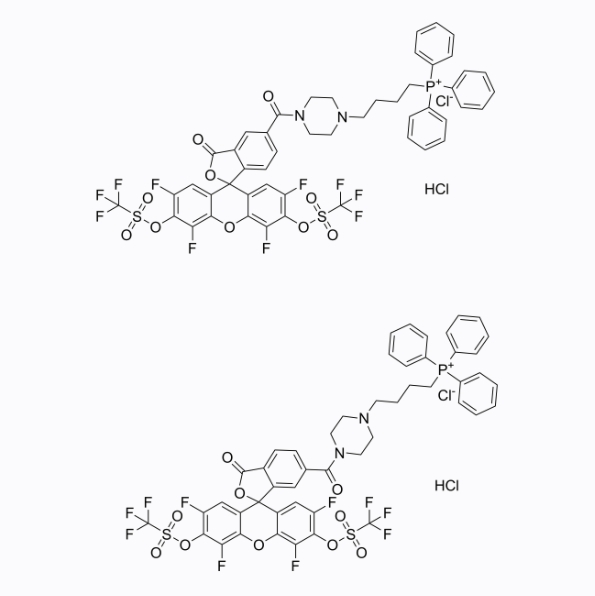
HKSOX-1m (5/6-mixture)
HKSOX-1m (5/6-mixture) is a reactive oxygen species fluorescent probe targeting mitochondria, with a maximum absorption/emission wavelength of 509/534nm.
-
GC26346
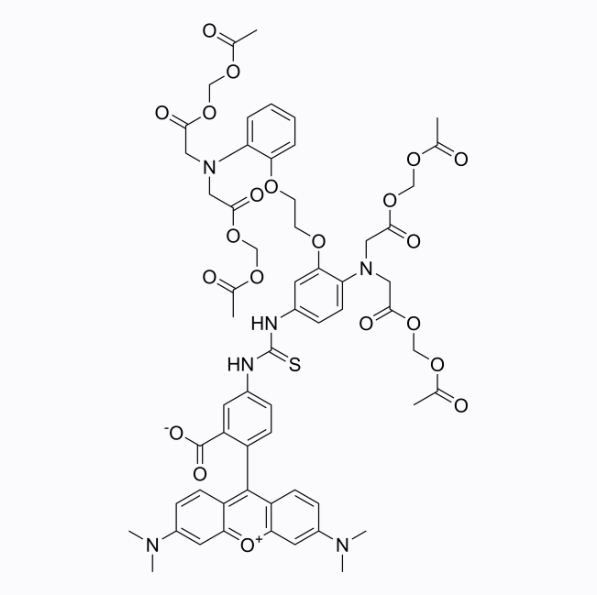
Calcium Orange AM
Calcium Orange AM is an intracellular calcium indicator in live cells. It can detect specific fluorescence after binding to free calcium. The maximum excitation wavelength and emission wavelength are 549/576 nm respectively.
-
GC26345
BODIPY 665/676 Lipid Peroxide Fluorescent Probe
BODIPY 665/676 is a lipophilic fluorescent probe with high affinity for binding free fatty acids and triglycerides, with a maximum absorption wavelength of long wavelength( λ Ex=665nm) and maximum fluorescence emission wavelength (Em=676nm)

-
GC26344
Nuclear and Cytoplasmic Protein Extraction Kit
Nuclear and Cytoplasmic Protein Extraction Kit provides a simple and convenient method for extracting nuclear and cytoplasmic proteins from cultured cells or fresh tissues.

-
GC26343
HRP-conjugated Streptavidin
Streptavidin is a biotin-binding protein derived from Streptomyces avidinii.

-
GC26342
Triflusulfuron-methyl

-
GC26341
Cupric hydroxide, AR

-
GC26340
2-(2-Chloro-4-(methylsulfonyl)-3-((2,2,2-trifluoroethoxy)methyl)benzoyl)cyclohexane-1,3-dione

-
GC26339
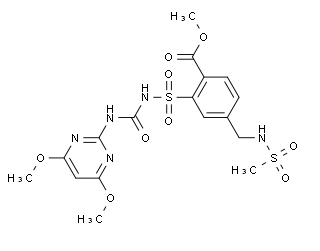
Mesosulfuron-Methyl
-
GC26338
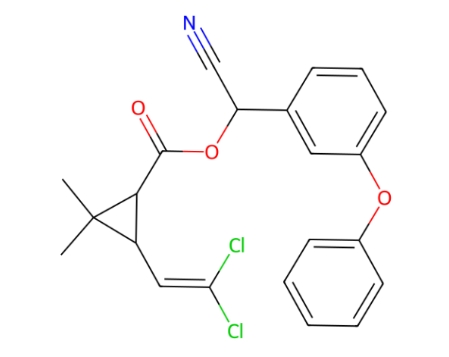
α-Cypermethrin
-
GC26337
REHMANNIAE RADIX PRAEPARATA

-
GC26335
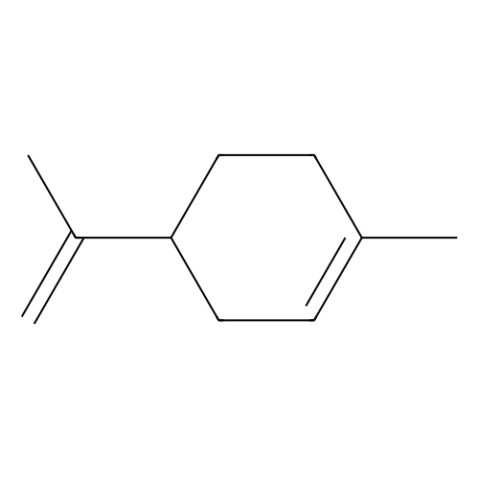
Dipentene
-
GC26334
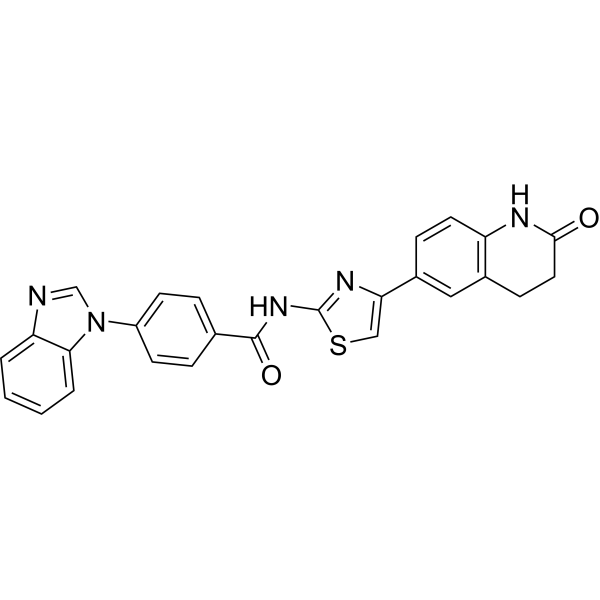
SRI 37892
SRI 37892 is a small molecule compound inhibitor of Frizzled protein 7 (Fzd7) with inhibitory activity against cancer cell proliferation (IC502μM).
-
GC26333
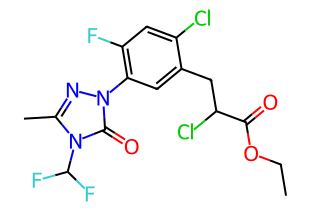
Carfentrazone-ethyl
-
GC26332
PVP (M.W.~ 58000)

-
GC26331
PVP (M.W.~ 40000)

-
GC26330
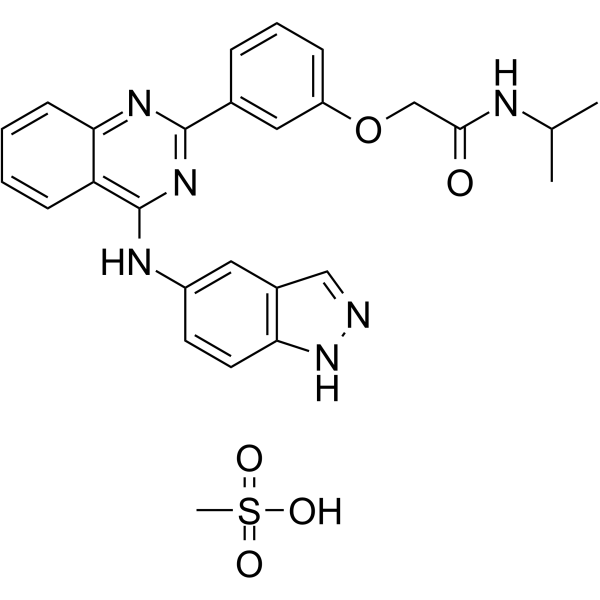
Belumosudil mesylate
Belumosudil mesylate (KD025 mesylate) is a selective inhibitor of ROCK2 with IC50 of 105 nM and 24 µM for ROCK2 and ROCK1, respectively.
-
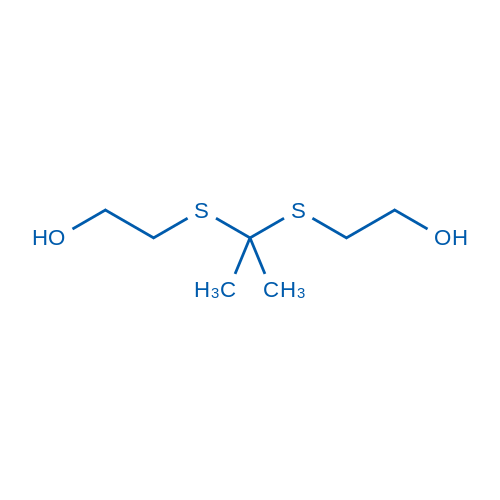
GC26329
2,2'-(Propane-2,2-diylbis(sulfanediyl))diethanol
-
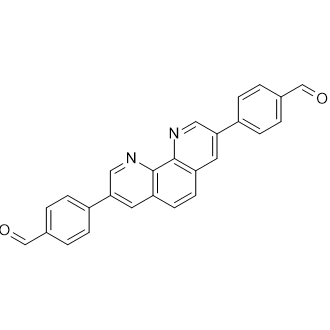
GC26336
4,4'-(1,10-Phenanthroline-3,8-diyl)dibenzaldehyde
-
GC26328
Etaracizumab
Etaracizumab (LM 609) is an αvβ3 integrin IgG1 monoclonal antibody.

-
GC26327
Cell-Tracker Red CMTPX
Cell Tracker fluorescent probes are excellent tools for monitoring cell movement, localization, proliferation, migration, chemotaxis, and invasion.
-
GC26326
1,3,4-TRIMETHYL-3-CYCLOHEXEN-1-CARBOXALDEHYDE

-
GC26325
Potassium nitrate-15N(99atom%)

-
GC26324
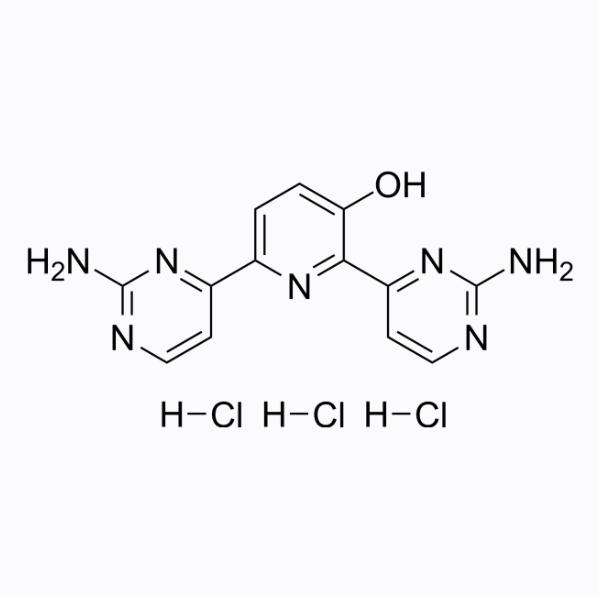
Avotaciclib trihydrochloride
Avotaciclib (BEY1107) trihydrochloride is an effective and orally active cyclin dependent kinase 1 (CDK1) inhibitor.
-
GC26323
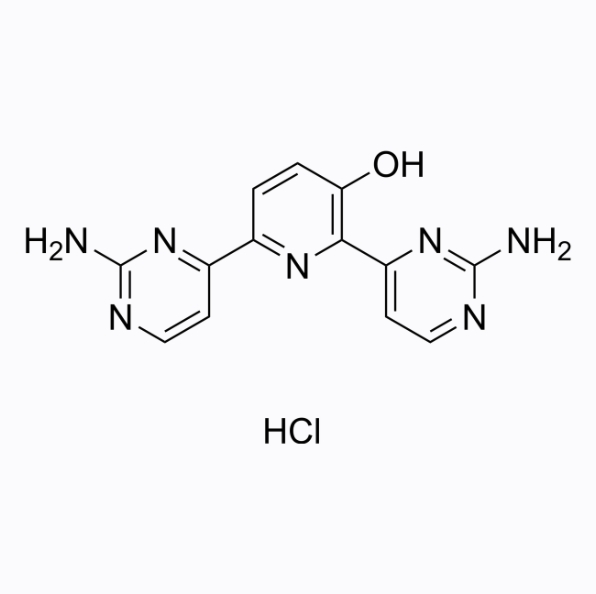
Avotaciclib hydrochloride
Avotaciclib hydrochloride is the hydrochloride form of CDK1 inhibitor Avotaciclib.
-
GC26322
Dalotuzumab
Dalotuzumab (MK-0646) is a recombinant humanized monoclonal antibody (IgG1 type) targeting IGF-1R. Dalotuzumab works by inhibiting IGF-1 and IGF-2 mediated tumor cell proliferation, IGF-1R autophosphorylation, and Akt phosphorylation.

-
GC26321
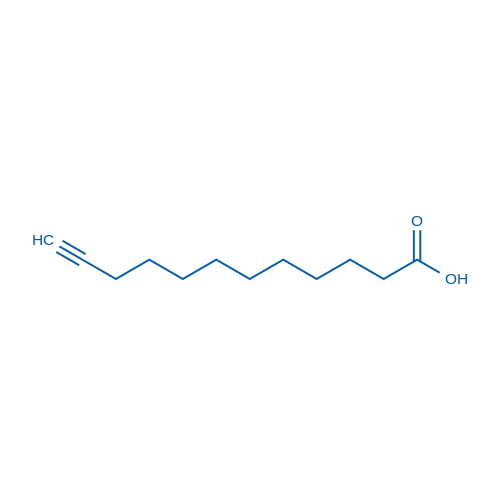
Dodec-11-ynoic acid
-
GC26320
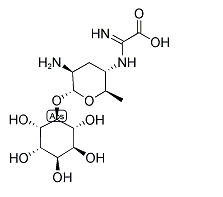
KASUGAMYCIN
Chunrimycin is an antibiotic that works by binding to the mRNA channel of the 30S subunit and inhibiting protein synthesis.
-
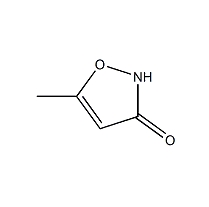
GC26318
Hymexazol
-
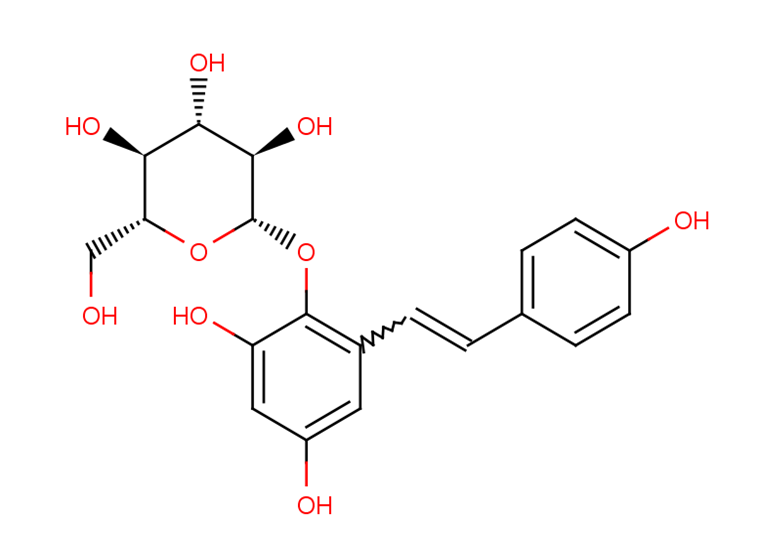
GC26317
Cassiaside B2
Cassiaside B2 is an inhibitor of protein tyrosine phosphatase 1B (PTP1B) and human monoamine oxidase A (hMAO-A).Cassiaside B2 is an agonist of the 5-HT2C receptor with anti-allergic activity.Cassiaside B2 is one of the active ingredients in the naphthylpyrone reference extract (NRE).
-
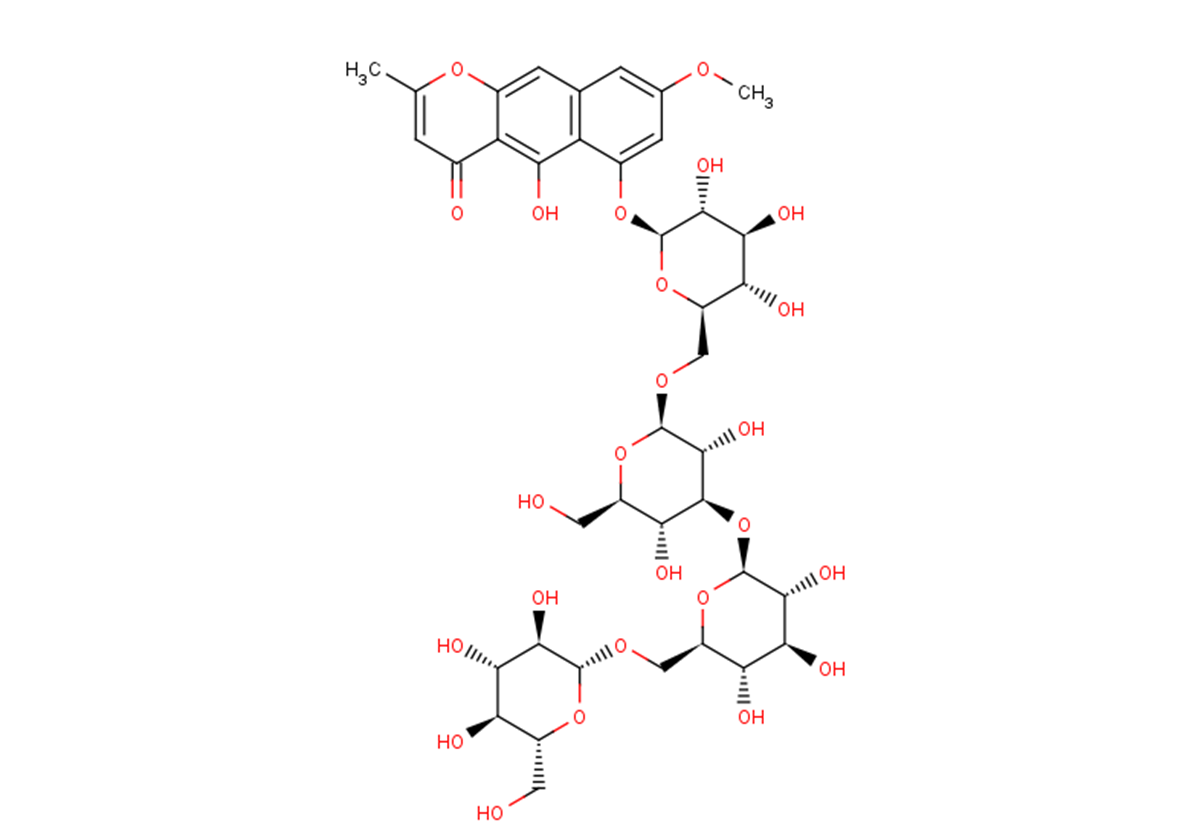
GC26316
Tetrahydroxystilbene-2-O-β-D-glucoside
Tetrahydroxysilbene-2-O- β- D-glucoside (EH-201) is a low molecular weight erythropoietin inducer.
-
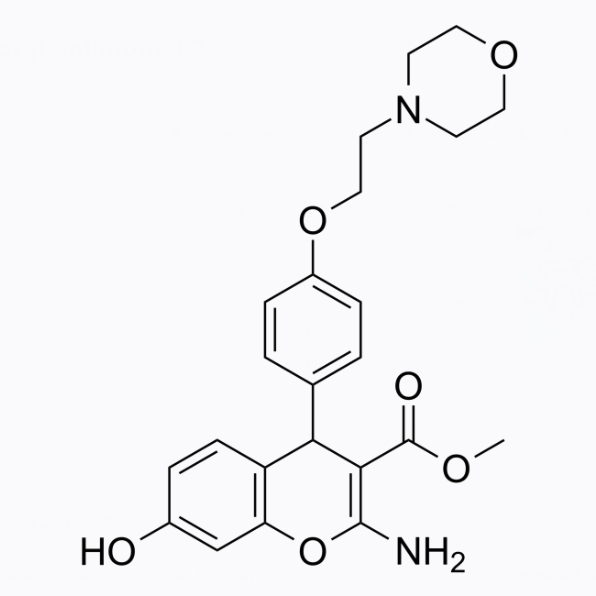
GC26315
Estrogen receptor β antagonist 2
Estrogen receiver β Antagonist 2 is an effective selective estrogen receptor β Estrogen receiver β (ER) β) Antagonists against Er α And Er β The IC50 value is 109.10, 0.63 µ M.
-
GD67968
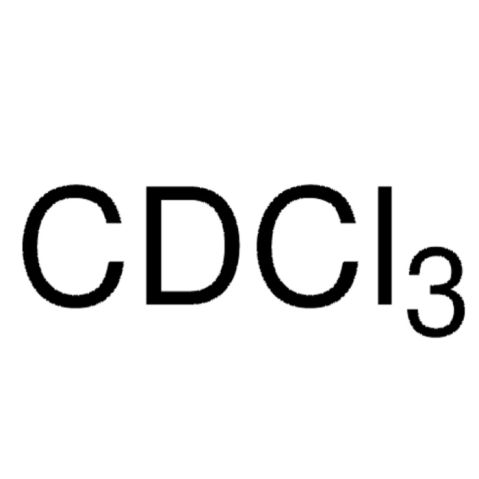
Chloroform-d
Chloroform-d (Deuterochloroform, CDCl3) is a deuterated derivative of chloroform.
-
GC26314
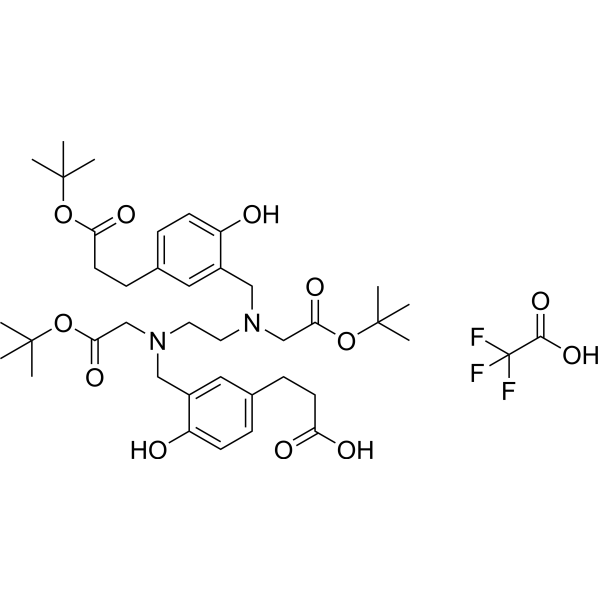
HBED-CC-tris(tert-butyl ester) TFA
HBED-CC-tris(tert-butyl ester) TFA, a HBED-CC derivative, is a bifunctional chelating agent.
-
GC26313
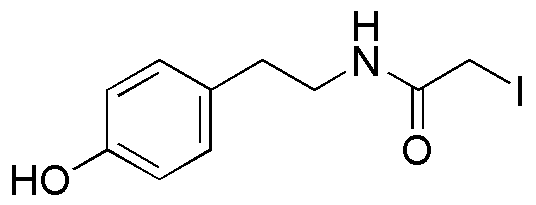
β-(4-Hydroxyphenyl)ethyl iodoacetamide
-
GC26312
Glycerol Jelly Mounting Medium
Glycerol Jelly Installing Medium is a water-based sealing solution prepared using the classic Kaiser method. However, it has been improved to no longer contain highly toxic phenol and can be used for conventional sealing of various slices, smears, etc.

-
GC26311
Lysing Enzymes
Lysozyme originates from Arthrobacter vinifera

-
GC26310
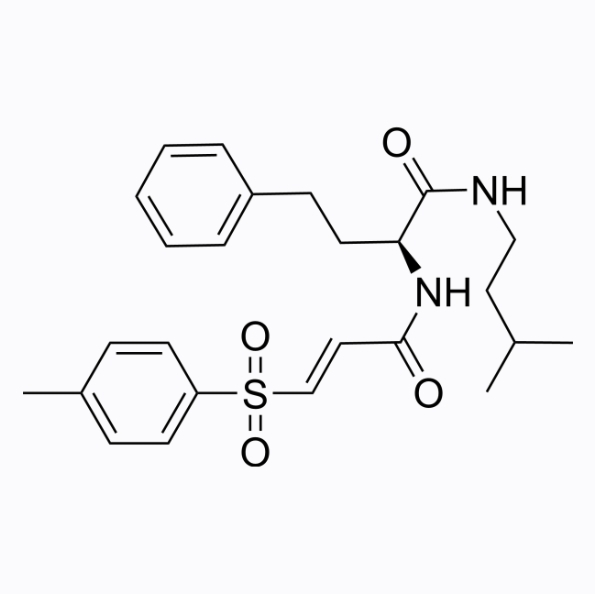
WRR139
WRR139 is a peptide vinyl sulfone that is involved in disease processes such as inflammation and cancer.
-
GC26309
Lactate Oxidase (≥20u/mg solid)

-
GC26308
Lactate Oxidase ( ≥80 units/mg solid)
Enzyme activity definition:One unit causes the formation of one micromole of hydrogen peroxide (half a micromole of quinoneimine dye) per minute under the conditions described below.

-
GC26307
1V-LSD (solution)
1V-LSD (solution)is an analytical reference material categorized as a lysergamide.
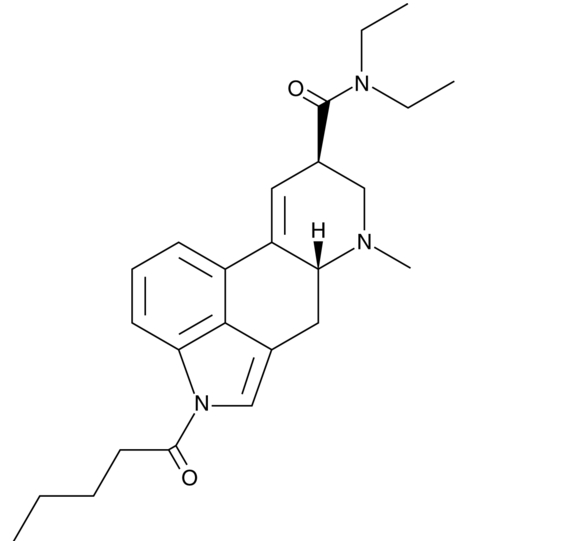
-
GC26306
PKH 67
PKH67 is a fluorescent cell junction dye with green fluorescence.

-
GC26305
NH2-UAMC1110 TFA
NH2-UAMC1110 TFA is a UAMC1110 derivative that can be used for the synthesis of FAPI-QS. UAMC1110 is a fibroblast activating protein (FAP) inhibitor.
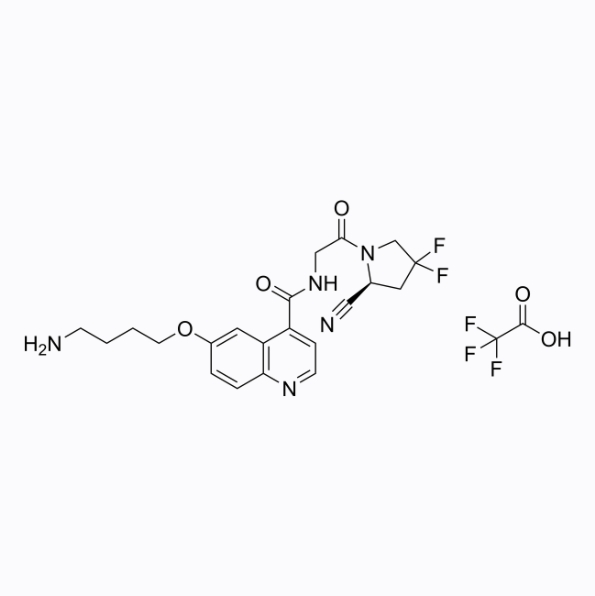
-
GC26304
3,4-Methylenedioxy-α-propylaminobutiophenone (hydrochloride) (exempt preparation)
3,4-Methylenedioxy-α-propylaminobutiophenone (hydrochloride) (exempt preparation) is an analytical reference standard categorized as a cathinone.
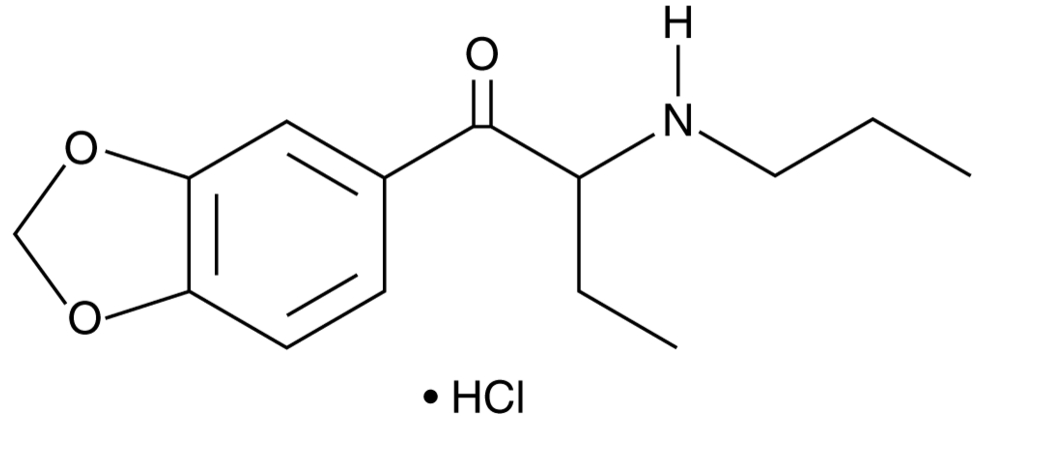
-
GC26303
[Tyr8] Bradykinin
[Tyr8] Bradykinin is a B2 kinin receptor agonist.
![[Tyr8] Bradykinin Chemical Structure [Tyr8] Bradykinin Chemical Structure](/media/struct/GC2/GC26303.png)
-
GC26302
Ni-NTA His-Tag Purification Agarose
Ni-NTA His-Tag Purification Agarose consists of nitrilotriacetic acid (NTA) chelator-activated agarose beads that subsequently charged with divalent nickel (Ni2+) ions by four coordination sites.

-
GC26301
Pd-PEPPSI-IPr
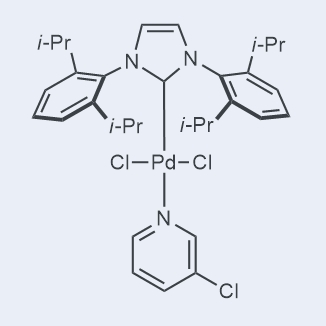
-
GC26300
Ammonium thiocyanate
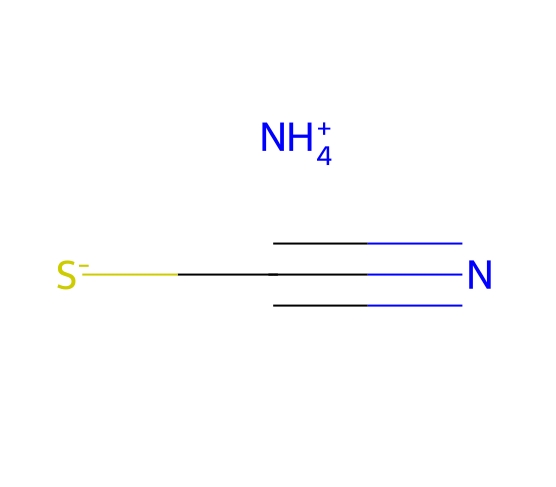
-
GC26299
Belantamab mafodotin

-
GC26298
Patritumab deruxtecan
Patritumab deruxtecan (HER3-DXd) is an antibody-drug conjugate (ADC) consisting of a fully human anti-HER3 IgG1 monoclonal antibody Patritumab attached to a topoisomerase I inhibitor payload via a tetrapeptide-based cleavable linker.

-
GC26297
ODN 1018 sodium
ODN 1018 sodium, an oligodeoxynucleotide, is a TLR-9 agonist.

-
GC26296
Squalene
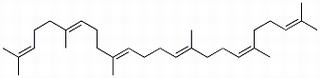
-
GC26295
Cis-N-Feruloyltyramine
Cis-N-Feruloyltyramine is a naturally occurring compound found in various plants that shows cytotoxicity against the P-388 cancer cell line. Cis-N-Feruloyltyramine is an inhibitor of in vitro prostaglandin (PG) synthesis.
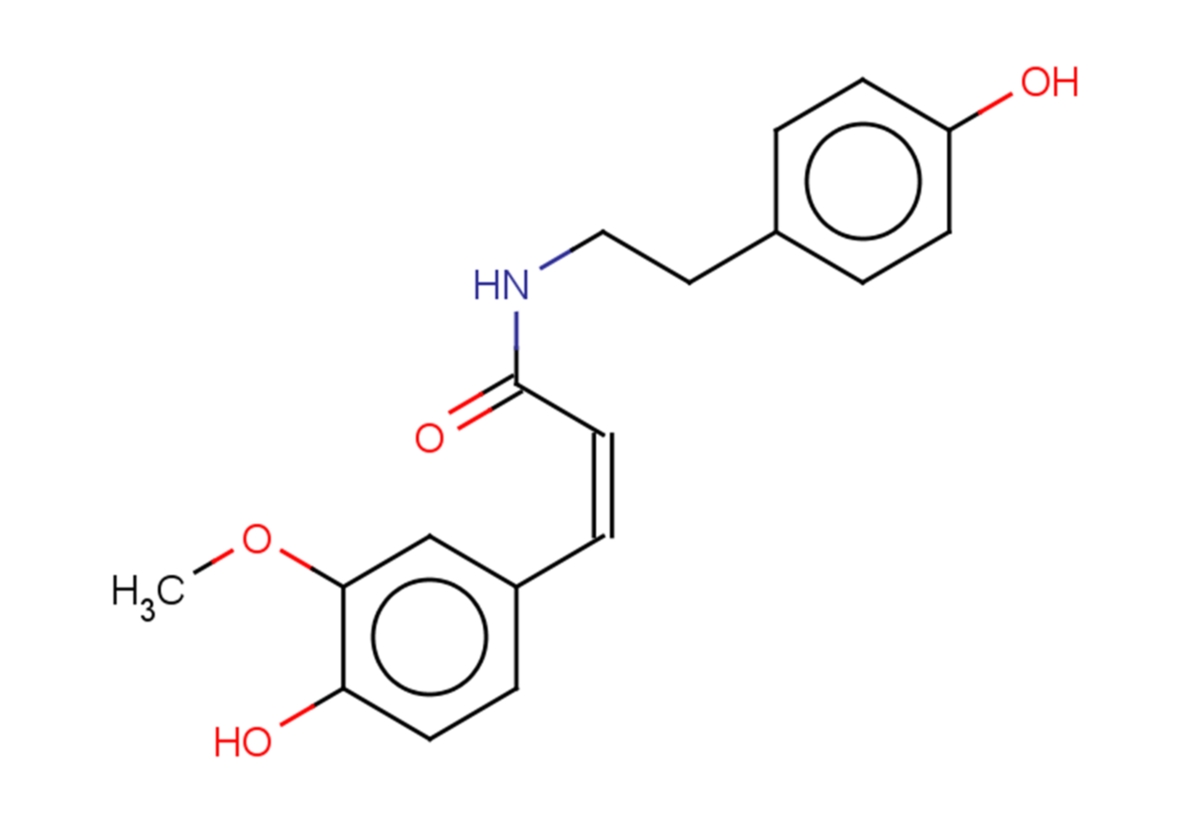
-
GC26294
Supelco 37 Component FAME Mix

-
GN10756
Ophiopogonin D
Extracted from Radix Liriopes;Store the product in sealed,cool and dry condition
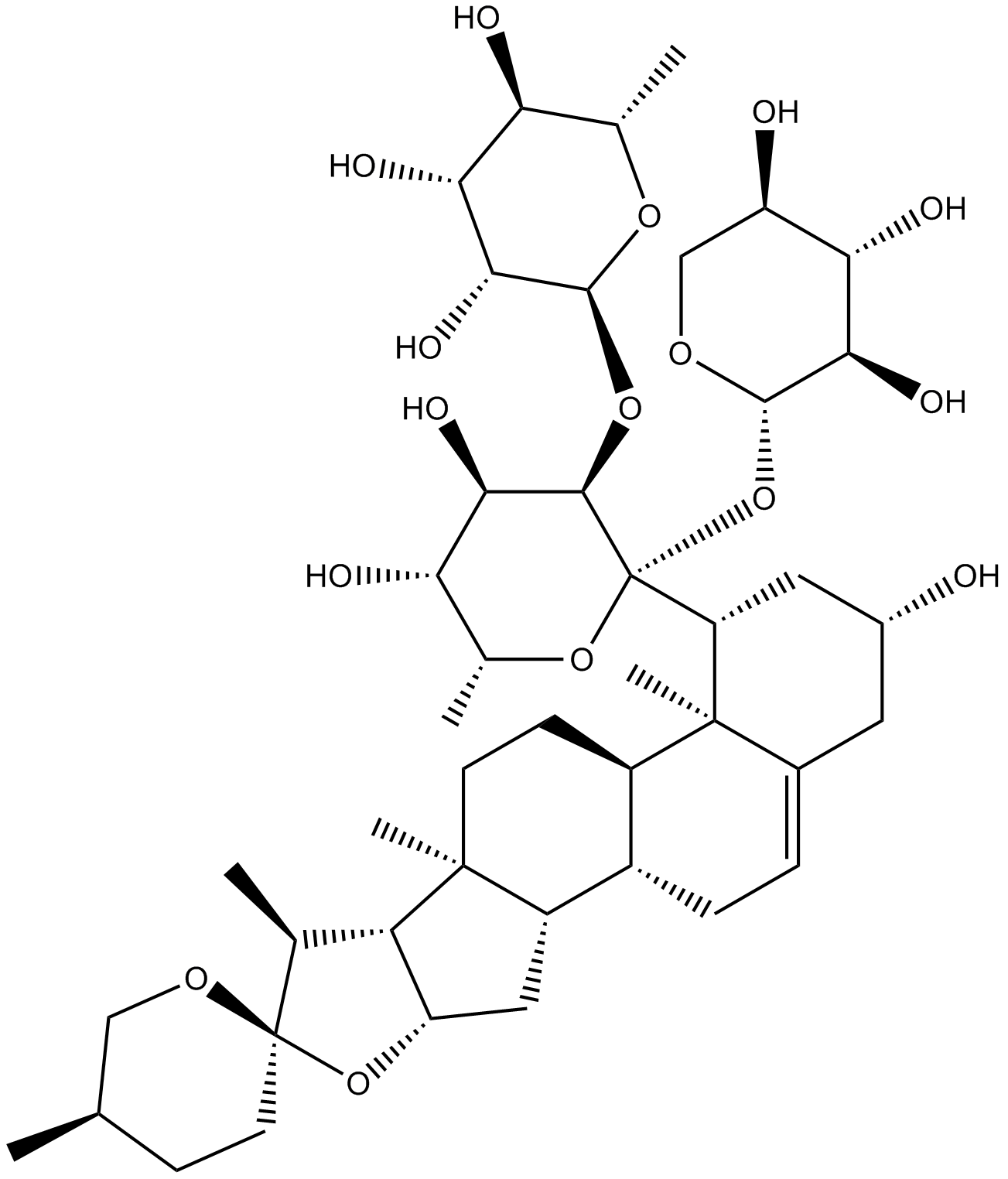
-
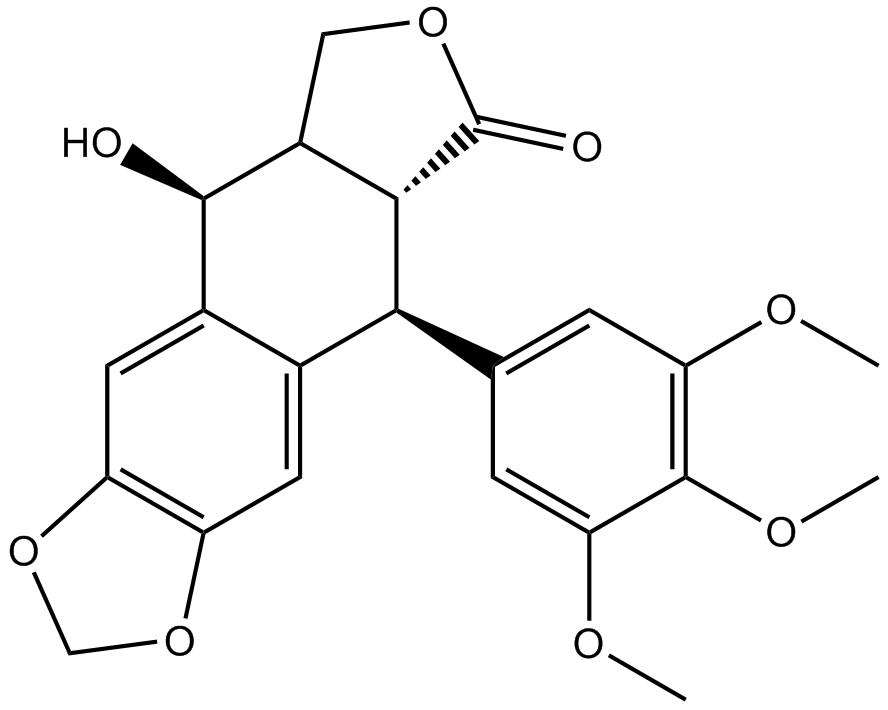
GN10416
Podophyllotoxin
A lignan with diverse biological activities
-
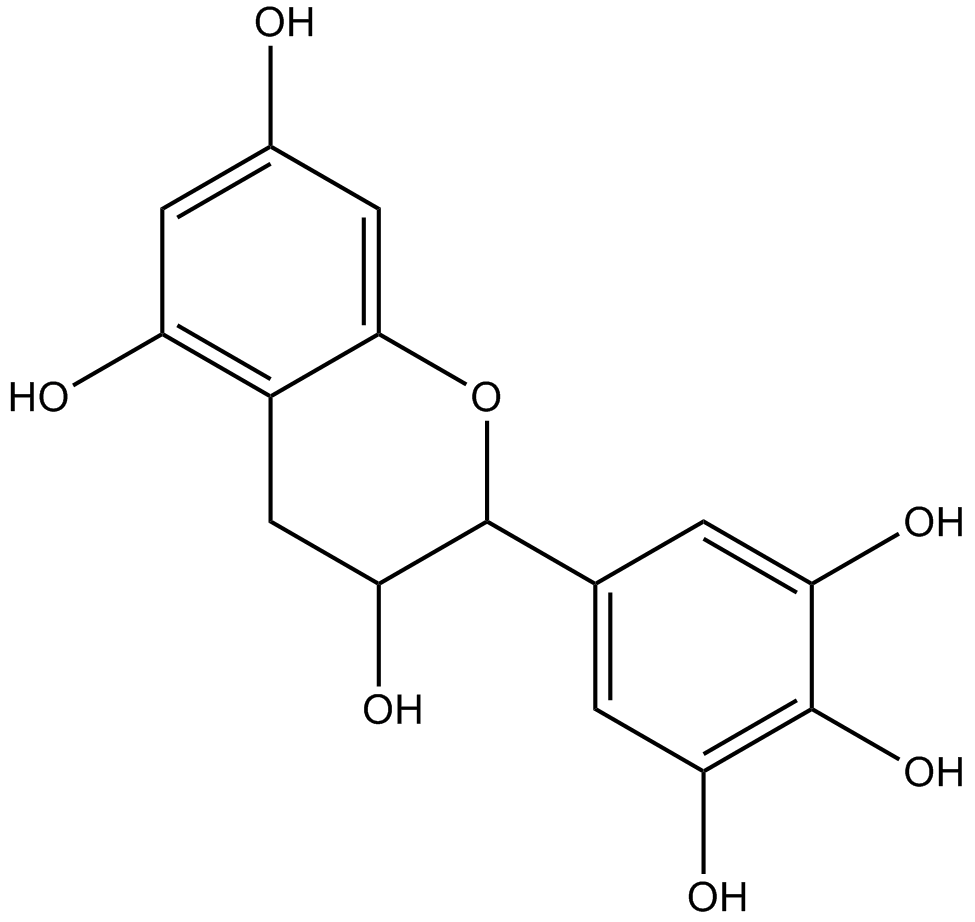
GN10366
Epigallocatechin
A green tea polyphenol
-
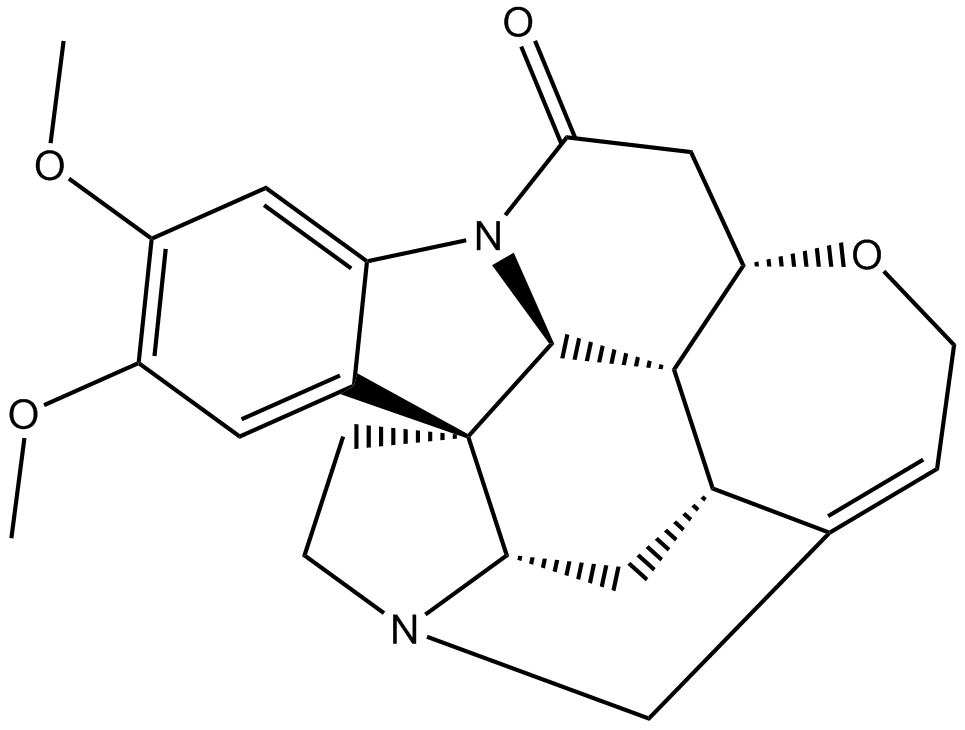
GN10235
Brucine
An alkaloid with diverse biological activities
-
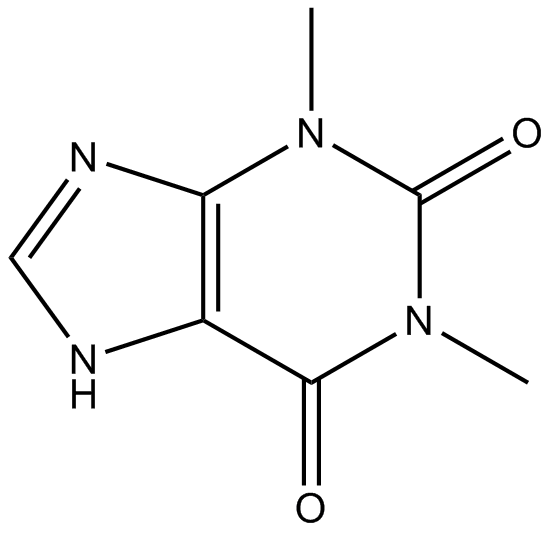
GN10659
Theophylline
A phosphodiesterase inhibitor and adenosine receptor antagonist
-
GC26170
9(R)-Hexahydrocannabinol (CRM)

-
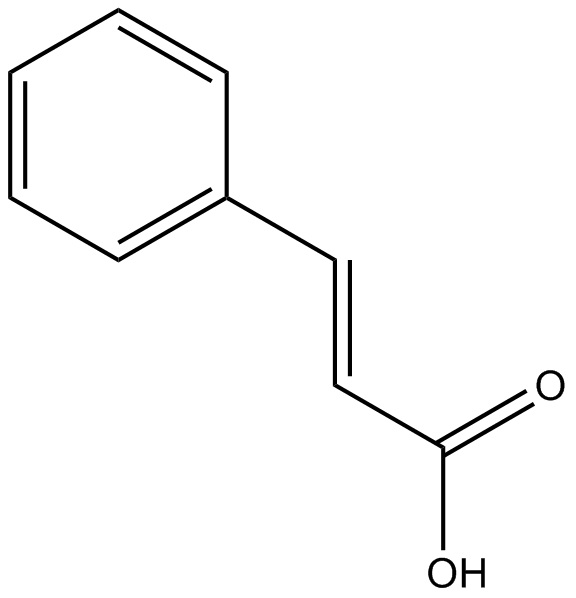
GN10190
Cinnamic acid
Unsaturated carboxylic acid
-
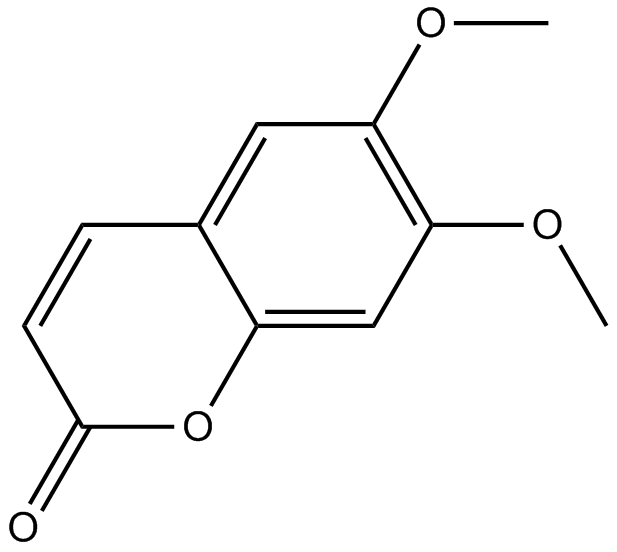
GN10816
Scoparone
A coumarin with diverse biological activities
-
GC26169
9(S)-Hexahydrocannabinol (CRM)
9(S)-Hexahydrocannabinol (CRM) is a certified reference material that is structurally similar to known phytocannabinoids.

-
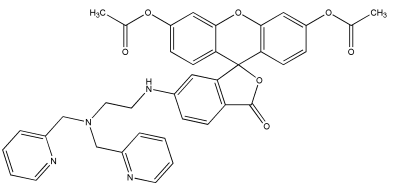
GC26167
ZnAF-2 DA,Cell Permeant
ZnAF-2 DA is a membrane-permeable derivative of the zinc ion fluorescent probe ZnAF-2.
-
GC26293
T4 Polynucleotide Kinase
T4 Polynucleotide Kinase, abbreviated as T4 PNK, is a type of polynucleotide 5 'hydroxy kinase,

-
GC26292
Bovine Serum Albumin Standard BSA standard (2 mg/mL)
BSA standard is a high-quality calibrator that is widely used to establish standard curves and calibration controls in total protein assay experiments.

-
GC26291
Lugol's iodine solution (Gram stain)
iodine concentration 0.33%

-
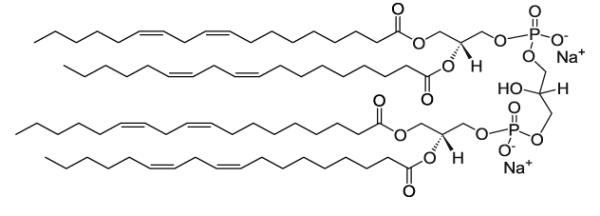
GC26290
Cardiolipin (Heart, Bovine) (sodium salt)
Cardiolipin is a unique phospholipid present in the inner mitochondrial membrane, which makes up to 20% of total lipids.
-
GC26289
Anti-Mouse PD-1 Antibody (RMP1-14)
Anti-Mouse PD-1 Antibody is an anti-mouse PD-1 IgG1 antibody inhibitor.

-
GC26288
Modified Giemsa Staining Solution(20X)
Modified Giemsa Staining Solution, also known as Wright Giemsa Staining Solution, is a staining solution used for staining samples such as cells, blood, bone marrow smears, or tissue sections.

-
GC26287
Factor Xa Protease
For Factor Xa, fusion protein cleavage is carried out at a w/w ratio of 1% the amount of fusion protein (e.g., 1 mg Factor Xa for a reaction containing 100 mg fusion protein).

-
GC26286
Cacodylic acid
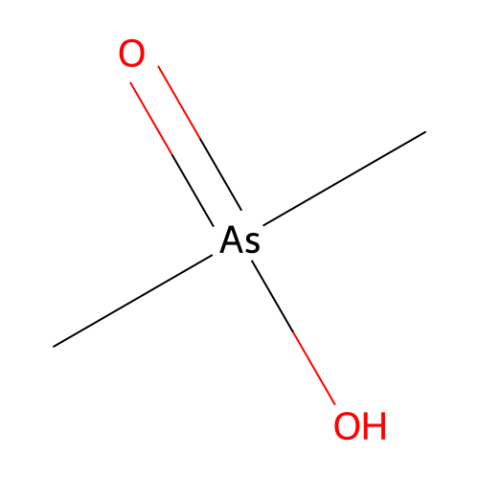
-
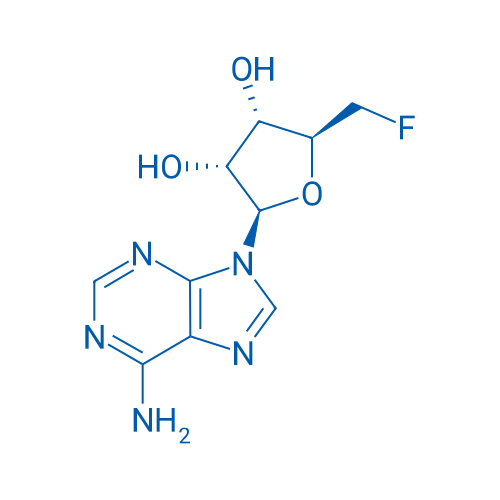
GC26285
(2R,3R,4S,5S)-2-(6-Amino-9H-purin-9-yl)-5-(fluoromethyl)tetrahydrofuran-3,4-diol
-
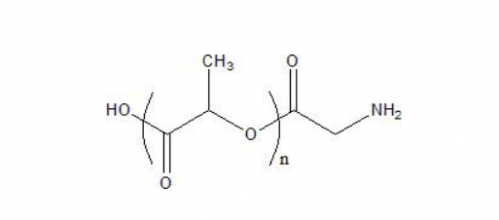
GC26284
PLLA-NH2 (M.Wt 50-60k)
PLA-NH2, Poly(D,L-lactide)-amine is a type of biodegradable polymer with an amine group on its surface.
-
GC26283
Antifade Mounting Medium with DAPI
Anti fluorescence mounting medium with DAPI is a sealing reagent that contains the blue fluorescent dye DAPI in the nucleus and can slow down fluorescence quenching.

-
GC26282
Agatolimod sodium
Agatolimod sodium (ODN 2006) is a B-class CpG ODN and a TLR9 agonist.

-
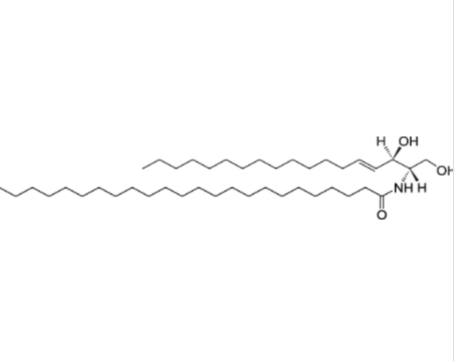
GC26281
C24 Ceramide (d18:1/24:0)
-
GK10027
Bradford Protein Quantification Kit
The Bradford Protein Quantification Kit is a kit for rapid determination of protein concentrations in the range of 0.025-2mg/ml.

-
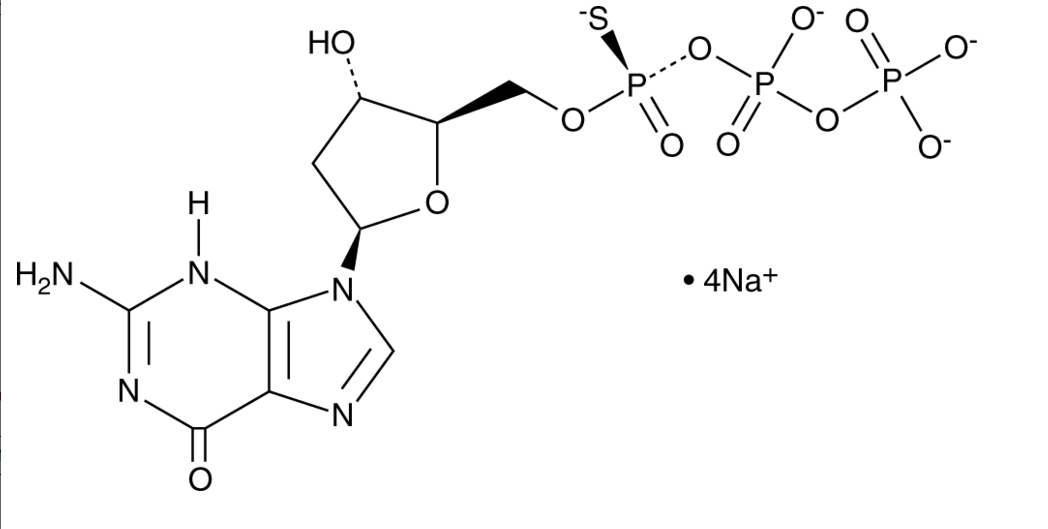
GC26280
Sp-2'-Deoxyguanosine-5'-O-(1-thiotriphosphate)
Sp-2'-Deoxyguanosine-5'-O-(1-thiotriphosphate) is a sulfur-containing isomer of the nucleoside triphosphate 2'-deoxyguanosine-5'-O-triphosphate.
-
GC26279
Tris-HCl Buffer,2.0M,pH7.5

-
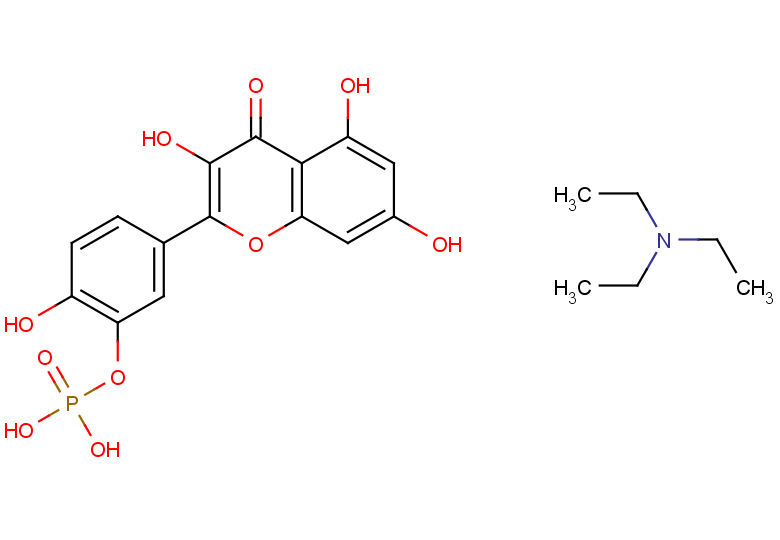
GC26278
Quercetin-3'-o-phosphate TEA
Quercetin-3'-o-phosphate TEA (Quercetin 3'-phosphate TEA) is an adenosine A receptor antagonist that can be used to prevent and treat metabolic disorders.
-
GC26277
Gamma glutamyltransferase
Gamma glutamyltransferase (EC 2.3.2.2) is a biochemical reagent that can be used as a biological material or organic compound for life science related research.

-
GC26276
Hoagland Nutrient Solution(without Phosphorus)
Hoagland Nutrient Solution(without Phosphorus)

-
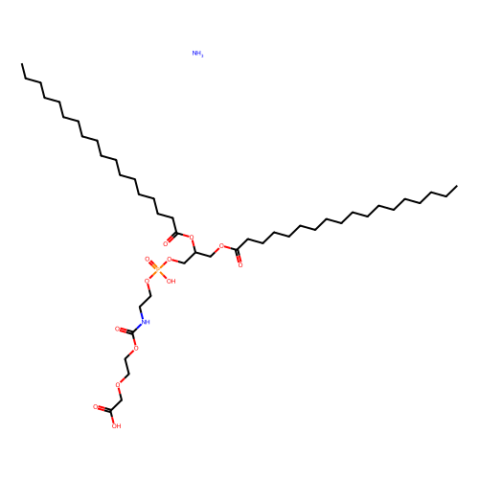
GC26275
DSPE-PEG2000-COOH
-
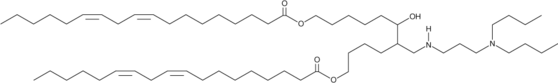
GC26274
IR-117-17
IR-117-17 is an ionizable cationic and biodegradable amino lipid.
-
GC26273
α-Amylase, heat-stable (BR, 40000u/g)
α-Amylase(heat-stable) (from Bacillus licheniformis), is widely used in various fermentation industries. This product is provided in solution form, with an enzyme activity of ≥500U/ml. The effective pH range of α-Amylase(heat-stable) is 5.5-8.0, with an optimal pH range of 5.8-6.5.

-
GC26272
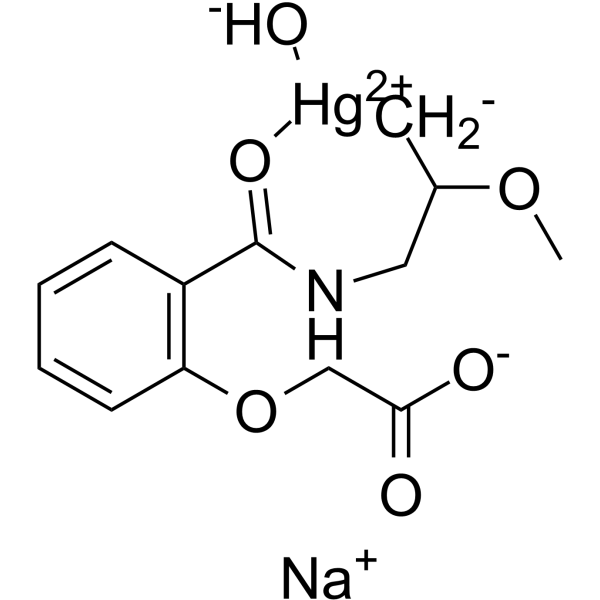
Mersalyl
Mersalyl (Salirgan) is a potent vascular endothelial growth factor (VEGF) and hypoxia-inducible factor 1 (HIF-1) inducer.
- GlpBio - Master of Small Molecules | Compounds - Peptides - Kits.
- Login | Register
- sales@glpbio.com
- (909) 407-4943
-
LanguageEN - English
-
Research Areas
-
Signaling Pathways
- Proteases
- Apoptosis
- Chromatin/Epigenetics
- Metabolism
- MAPK Signaling
- Tyrosine Kinase
- DNA Damage/DNA Repair
- PI3K/Akt/mTOR Signaling
- Microbiology & Virology
- Cell Cycle/Checkpoint
- Ubiquitination/ Proteasome
- JAK/STAT Signaling
- TGF-β / Smad Signaling
- Angiogenesis
- GPCR/G protein
- Stem Cell
- Cancer Biology
- Endocrinology and Hormones
- Immunology/Inflammation
- Neuroscience
- Membrane Transporter/Ion Channel
- Other Signal Transduction
- Product Type
-
Signaling Pathways
- Screening Library
- Kits
- Tools
- Ordering
- About us
- Contact us
- Blog


