Angiogenesis
Products for Angiogenesis
- Cat.No. Product Name Information
-
GC11753
P11
P11 is an antagonist of the integrin αvβ3-vitronectin interaction, with an IC50 of 1.74 pg/mL (2.414 pM).

-
GC11951
PAR 4 (1-6)
GYPGQV
PAR4 agonist

-
GC62101
PAR-2-IN-1
PAR-2-IN-1 is a protease-activated receptor-2 (PAR2) signaling pathway inhibitor with anti-inflammatory and anticancer effects.
-
GA23350
PAR-3 (1-6) amide (human)
PAR-3 (1-6) amide (human) is a proteinase-activated receptor (PAR-3) agonist peptide.
-
GC33599
PAR-4 Agonist Peptide, amide TFA (PAR-4-AP (TFA))
PAR-4-AP TFA; AY-NH2 TFA
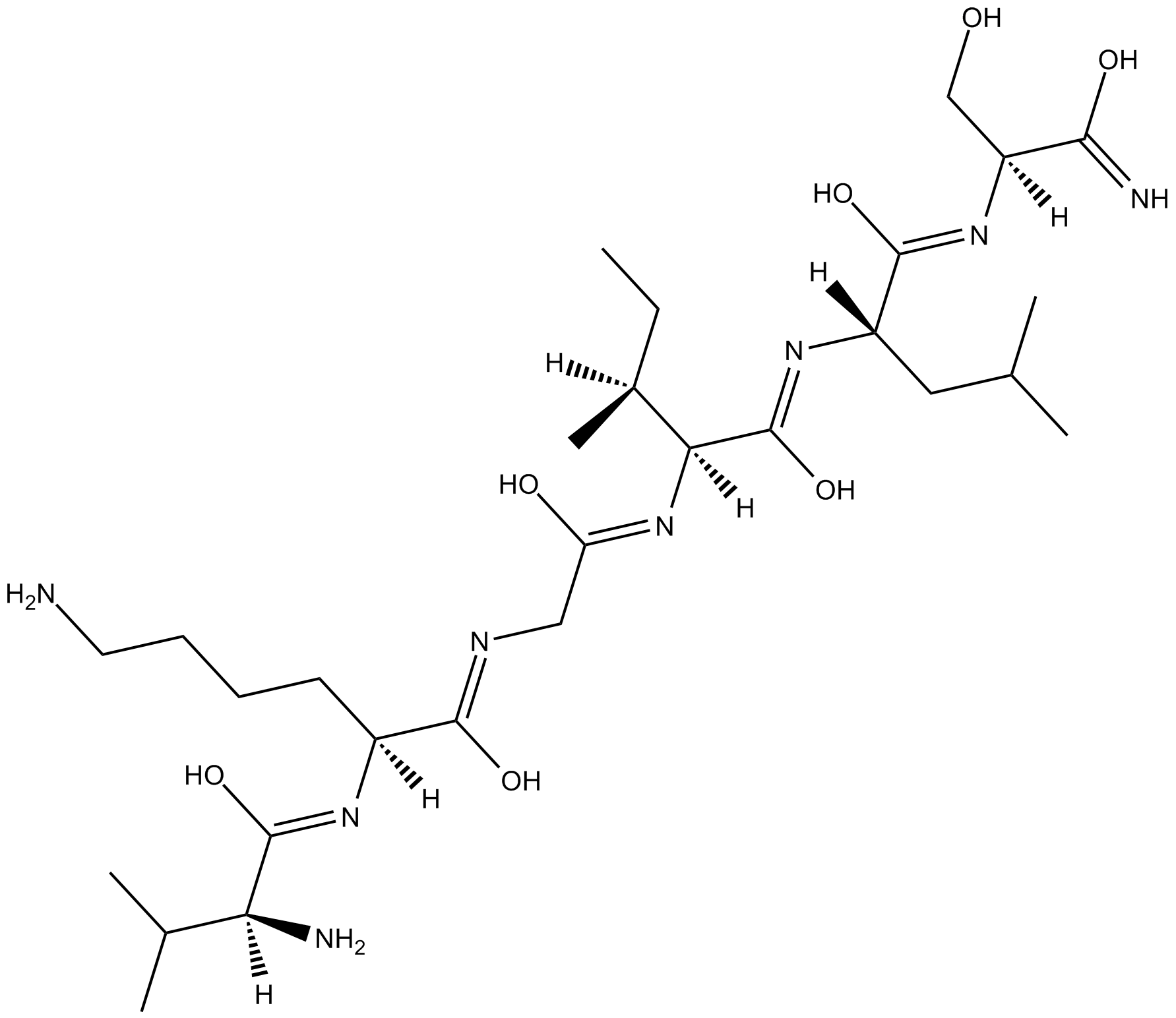
PAR-4 Agonist Peptide, amide TFA (PAR-4-AP (TFA)) (PAR-4-AP TFA; AY-NH2 TFA) is a proteinase-activated receptor-4 (PAR-4) agonist, which has no effect on either PAR-1 or PAR-2 and whose effects are blocked by a PAR-4 antagonist.
-
GC15817
PAR-4 Agonist Peptide, amide (AY-NH2)
PAR-4-AP; AY-NH2
PAR-4 Agonist Peptide, amide(AY-NH2) (PAR-4-AP; AY-NH2) is a proteinase-activated receptor-4 (PAR-4) agonist, which has no effect on either PAR-1 or PAR-2 and whose effects are blocked by a PAR-4 antagonist.
-
GC38134
Parmodulin 2
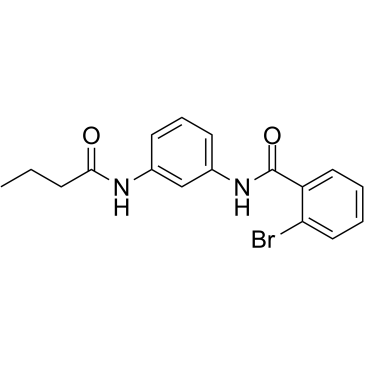
Reversible inhibitor of PAR1mediated platelet activation
-
GC69663
Parstatin(mouse) TFA
Parstatin (mouse) TFA is a peptide agonist of the PAR-1 thrombin receptor with cell-penetrating properties and an effective inhibitor of angiogenesis.

-
GC40085
Pazopanib-d6
Pazopanib-d6 is intended for use as an internal standard for the quantification of pazopanib by GC- or LC-MS.

-
GC36861
PCI 29732
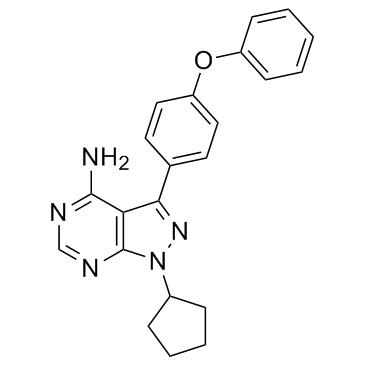
A multi-kinase inhibitor
-
GC13812
PCI-32765 (Ibrutinib)
Imbruvica, PCI 32765
PCI-32765 (Ibrutinib) is an irreversible selective inhibitor of Bruton s tyrosine kinase (BTK) that selectively targets its kinase domain and modulates BTK downstream signaling by reducing its phosphorylating capacity with IC50 of 0.5 nM in cell-free assays.
-
GC12921
PCI-32765 Racemate
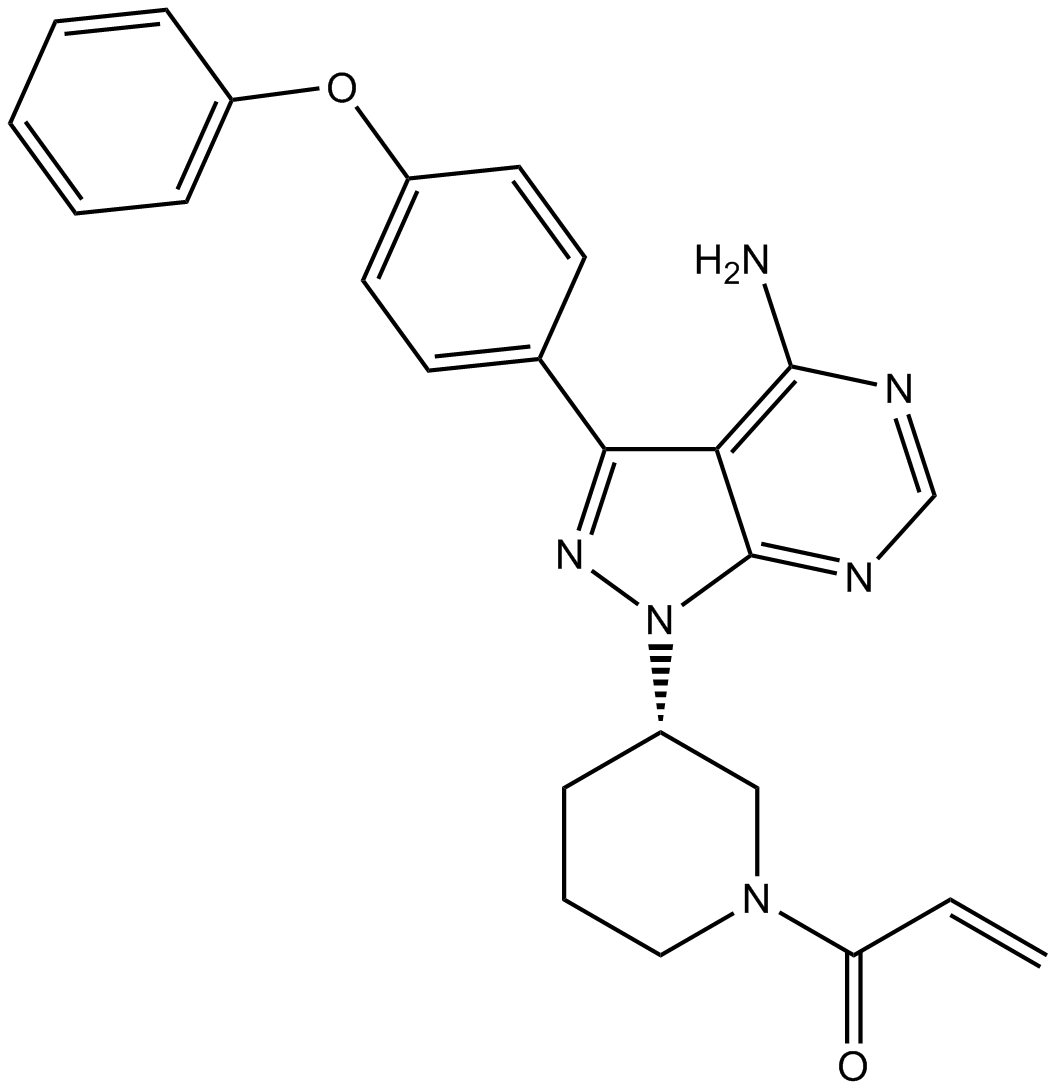
PCI-32765 Racemate (PCI-32765 Racemate) is the racemate of Ibrutinib. Ibrutinib is a selective, irreversible Btk inhibitor with IC50 value of 0.5 nM.
-
GC30155
PCI-33380
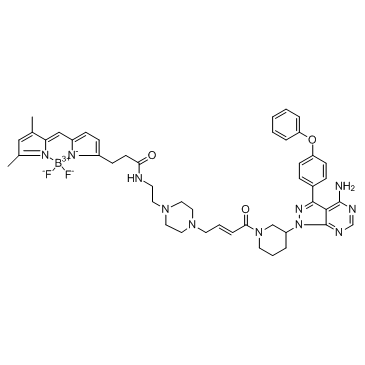
PCI-33380 is an irreversible and selective Bruton's Tyrosine Kinase (BTK) inhibitor (fluorescent probe).
-
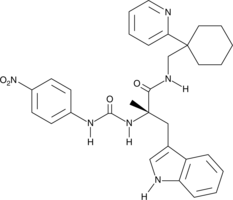
GC44584
PD 168368
PD 168368 is a competitive antagonist of neuromedin B (NMB) receptors (Kis = 15-45 nM for rat and human receptors expressed in various cell lines).
-
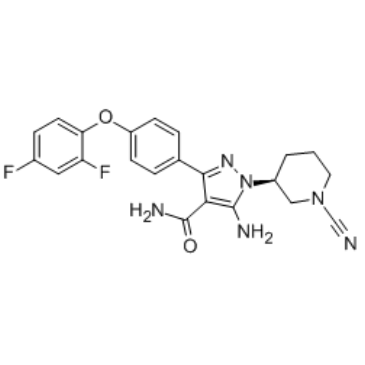
GC34709
PF-06250112
PF-06250112 is a potent, highly selective, orally bioavailable BTK inhibitor with an IC50 of 0.5 nM, shows inhibitory effect toward BMX nonreceptor tyrosine kinase and TEC with IC50s of 0.9 nM and 1.2 nM, respectively.
-
GC62515
Pirtobrutinib
LOXO-305, LY3527727
Pirtobrutinib (LOXO-305), a highly selective and non-covalent next generation BTK inhibitor, inhibits diverse BTK C481 substitution mutations. Pirtobrutinib causes regression of BTK-dependent lymphoma tumors in mouse xenograft models. Pirtobrutinib is also more than 300-fold selective for BTK versus 370 other kinases tested and shows no significant inhibition of non-kinase off-targets at 1 μM.
-
GC10235
Plinabulin (NPI-2358)
NPI-2358
Plinabulin (NPI-2358) (NPI-2358) is a vascular disrupting agen (VDA) against tubulin-depolymerizing with an IC50 of 9.8 nM against HT-29 cells. Plinabulin (NPI-2358) binds the colchicine binding site of β-tubulin preventing polymerization and has potent inhibitory to tumor cells.
-
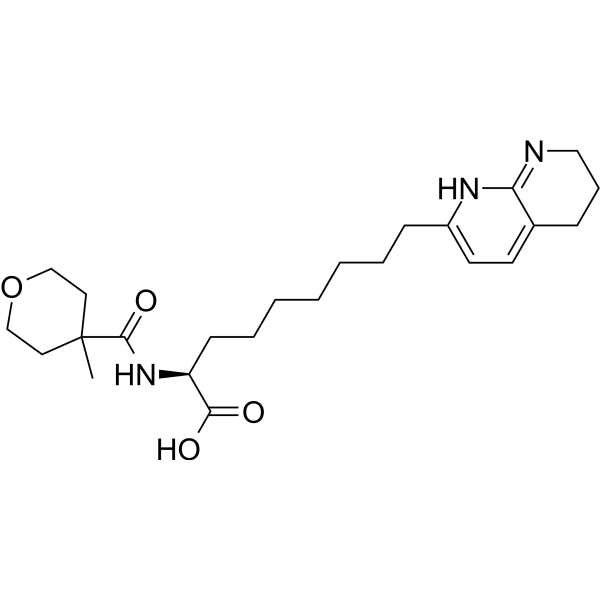
GC67708
PLN-1474
-
GC47965
Pomalidomide-d5
An internal standard for the quantification of pomalidomide

-
GC47966
PPACK (trifluoroacetate salt)
Pebac, DPhenylalanylprolylarginyl Chloromethyl Ketone
An antithrombin peptide
-
GC31339
PRN1008
PRN1008
PRN1008 (PRN1008) is a reversible covalent, selective and oral active inhibitor of Bruton's Tyrosine Kinase (BTK), with an IC50 of 1.3 nM.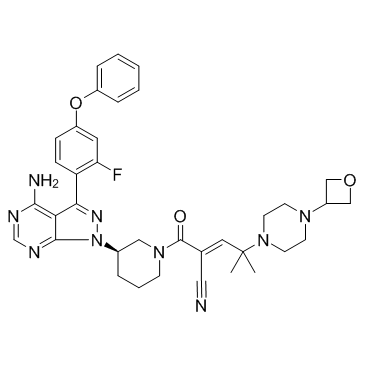
-
GC47980
Propionyl-L-carnitine-d3 (chloride)
C3:0 Carnitine-d3, CAR 3:0-d3, L-Carnitine propionyl ester-d3, Levocarnitine propionate-d3, L-Propionylcarnitine-d3
An internal standard for the quantification of propionyl-L-carnitine
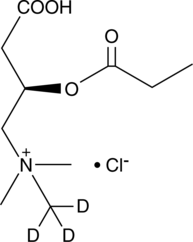
-
GC69768
Protease-Activated Receptor-1, PAR-1 Agonist
Protease-Activated Receptor-1, PAR-1 Agonist is a selective peptide that activates Protease-Activated Receptor-1 (PAR-1), which corresponds to the PAR1 ligand and selectively mimics the action of thrombin through this receptor.

-
GC62469
Protease-Activated Receptor-1, PAR-1 Agonist TFA
Protease-Activated Receptor-1, PAR-1 Agonist TFA is a selective proteinase-activated receptor1 (PAR-1) agonist peptide. Protease-Activated Receptor-1, PAR-1 Agonist TFA corresponds to PAR1 tethered ligand and which can selectively mimic theactions of thrombin via this receptor.
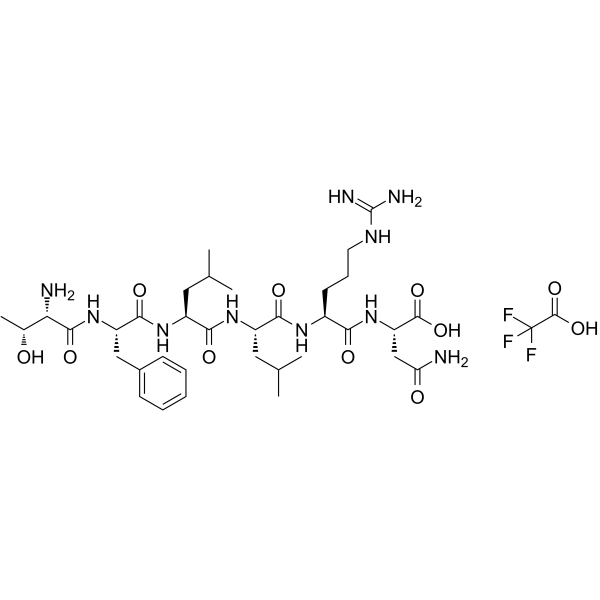
-
GC61456
Protease-Activated Receptor-3 (PAR-3) (1-6), human TFA
Protease-Activated Receptor-3 (PAR-3) (1-6), human TFA is a proteinase-activated receptor (PAR-3) agonist peptide.
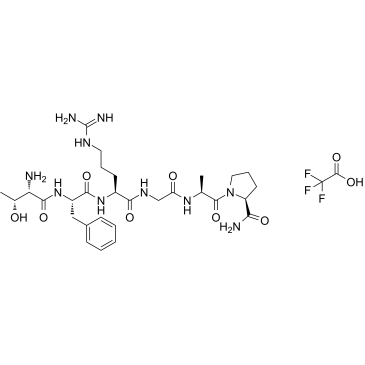
-
GC33429
Protease-Activated Receptor-4
Protease-Activated Receptor-4 is the agonist of proteinase-activated receptor-4 (PAR4).

-
GC49873
Protectin D1 methyl ester
Neuroprotectin D1 methyl ester, NPD1 methyl ester, PD1 methyl ester
An esterified form of protectin D1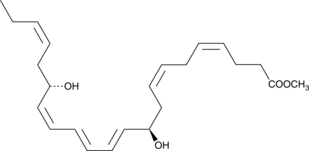
-
GC32680
PT-2385
PT-2385 is a selective HIF-2α inhibitor with a Ki of less than 50 nM.

-
GC37034
PT2977
PT2977; MK-6482
PT2977 (PT2977) is an orally active and selective HIF-2α inhibitor with an IC50 of 9 nM. PT2977, as a second-generation HIF-2α inhibitor, increases potency and improves pharmacokinetic profile. PT2977 is a potential treatment for clear cell renal cell carcinoma (ccRCC).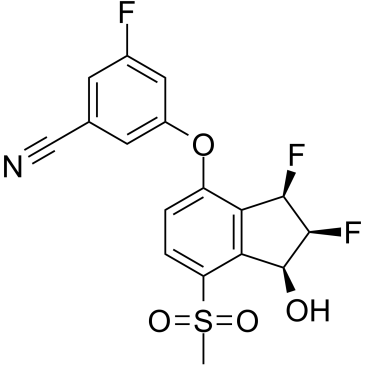
-
GC60311
PTUPB
PTUPB is a novel dual COX-2 and sEH inhibitor with IC50 values of 1.26 nM and 0.9 μM, respectively.

-
GC11031
PX-478 2HCl
PX-478 is a selective inhibitor that suppresses constitutive and hypoxia-induced HIF-1α levels.

-
GC37048
QL47
QL47, a broad-spectrum antiviral agent, inhibits dengue virus and other RNA viruses.
-
GC49221
QLT0267
An ILK inhibitor
-
GC44800
Quininib
A CysLT1 and CysLT2 receptor antagonist

-
GC37066
Rac-PT2399
A potent and specific HIF-2α inhibitor

-
GC40213
Regorafenib-13C-d3
Regorafenib-13C-d3 is intended for use as an internal standard for the quantification of regorafenib by GC- or LC-MS.

-
GC38375
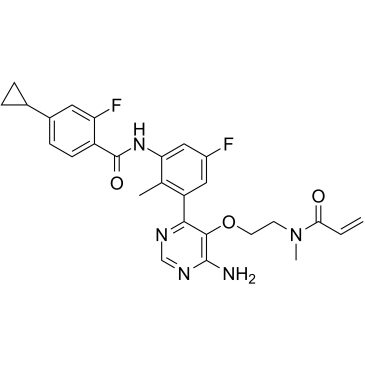
Remibrutinib
Remibrutinib, is a potent and orally active bruton tyrosine kinase (BTK) inhibitor with an IC50 value of 1 nM.
-
GC10184
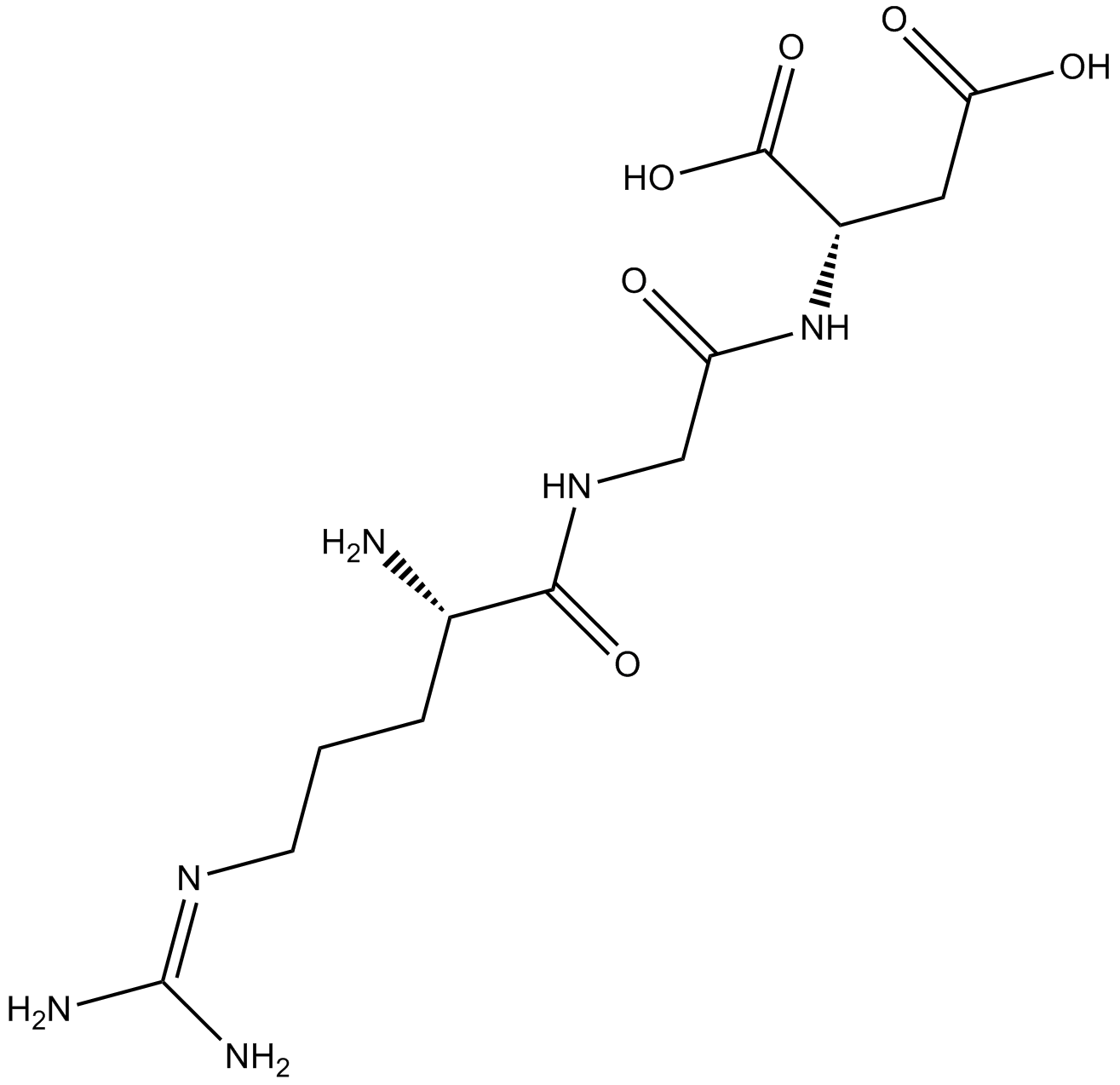
RGD (Arg-Gly-Asp) Peptides
RGD (Arg-Gly-Asp) Peptides is a tripeptide that effectively triggers cell adhesion, addresses certain cell lines and elicits specific cell responses; binds to integrins.
-
GC37524
RGD peptide (GRGDNP) TFA

-
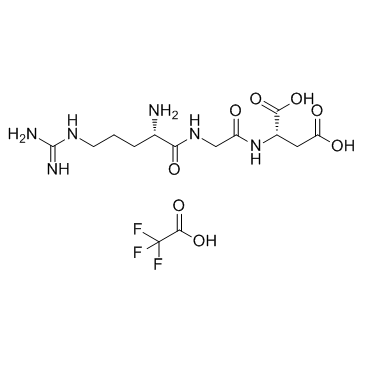
GC34402
RGD Trifluoroacetate
RGD Trifluoroacetate is a tripeptide that effectively triggers cell adhesion, addresses certain cell lines and elicits specific cell responses; RGD Trifluoroacetate binds to integrins.
-
GC16385
RGDS peptide
Fibronectin Inhibitor, HArgGlyAspSerOH
RGDS peptide is an integrin binding sequence that inhibits integrin receptor function.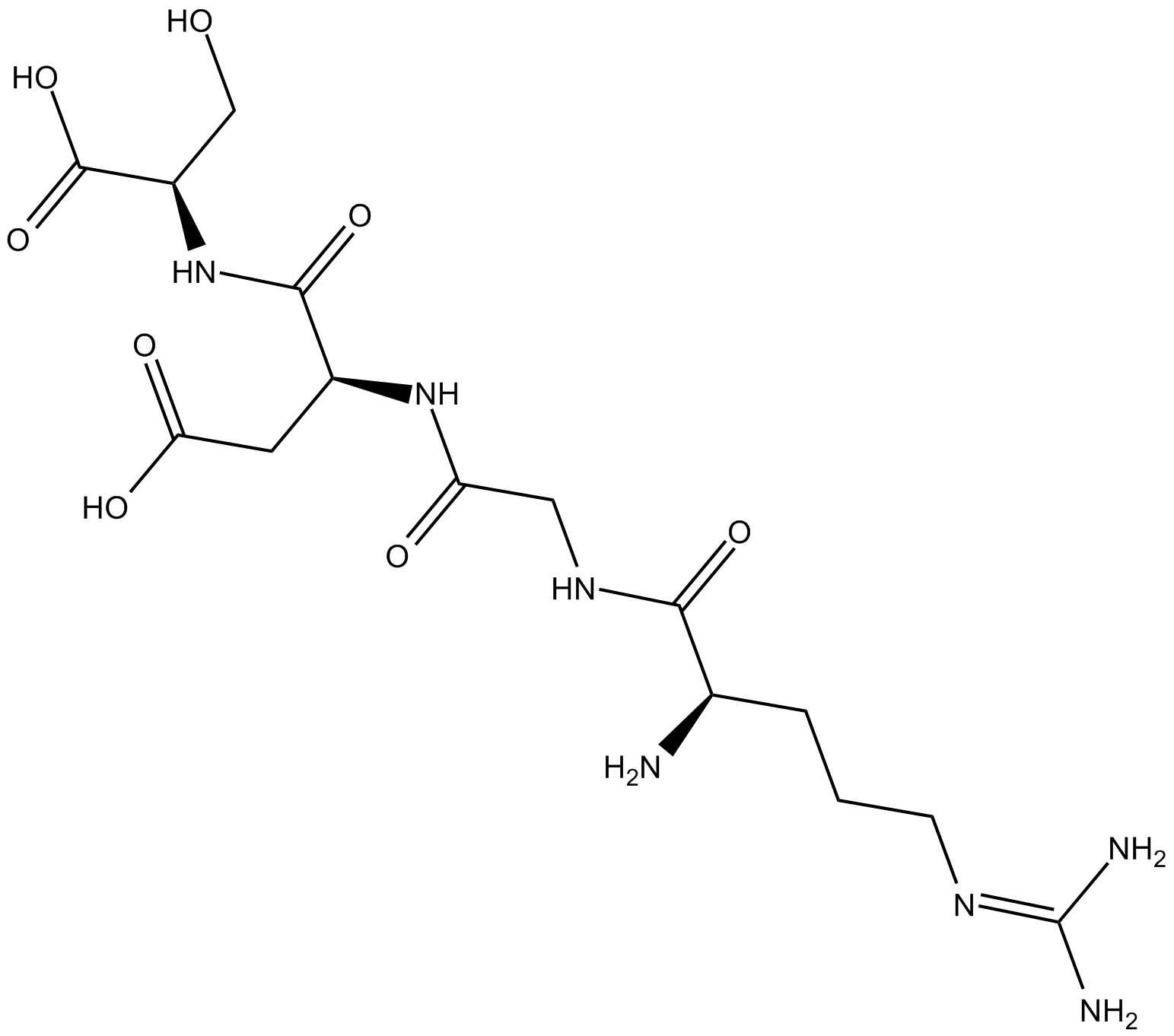
-
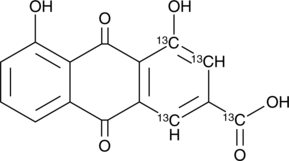
GC45798
Rhein-13C4
Rheic Acid-13C4
An internal standard for the quantification of rhein
-
GC37541
Risuteganib
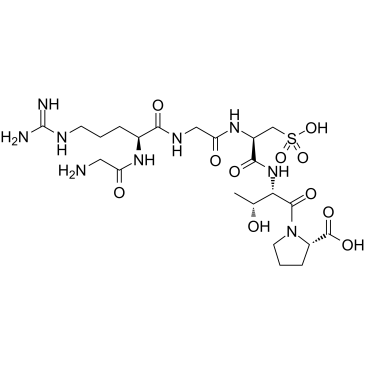
-
GC17991
RLLFT-NH2
Protease-activated receptor agonist

-
GC11230
RN486
Btk inhibitor,potent and selective
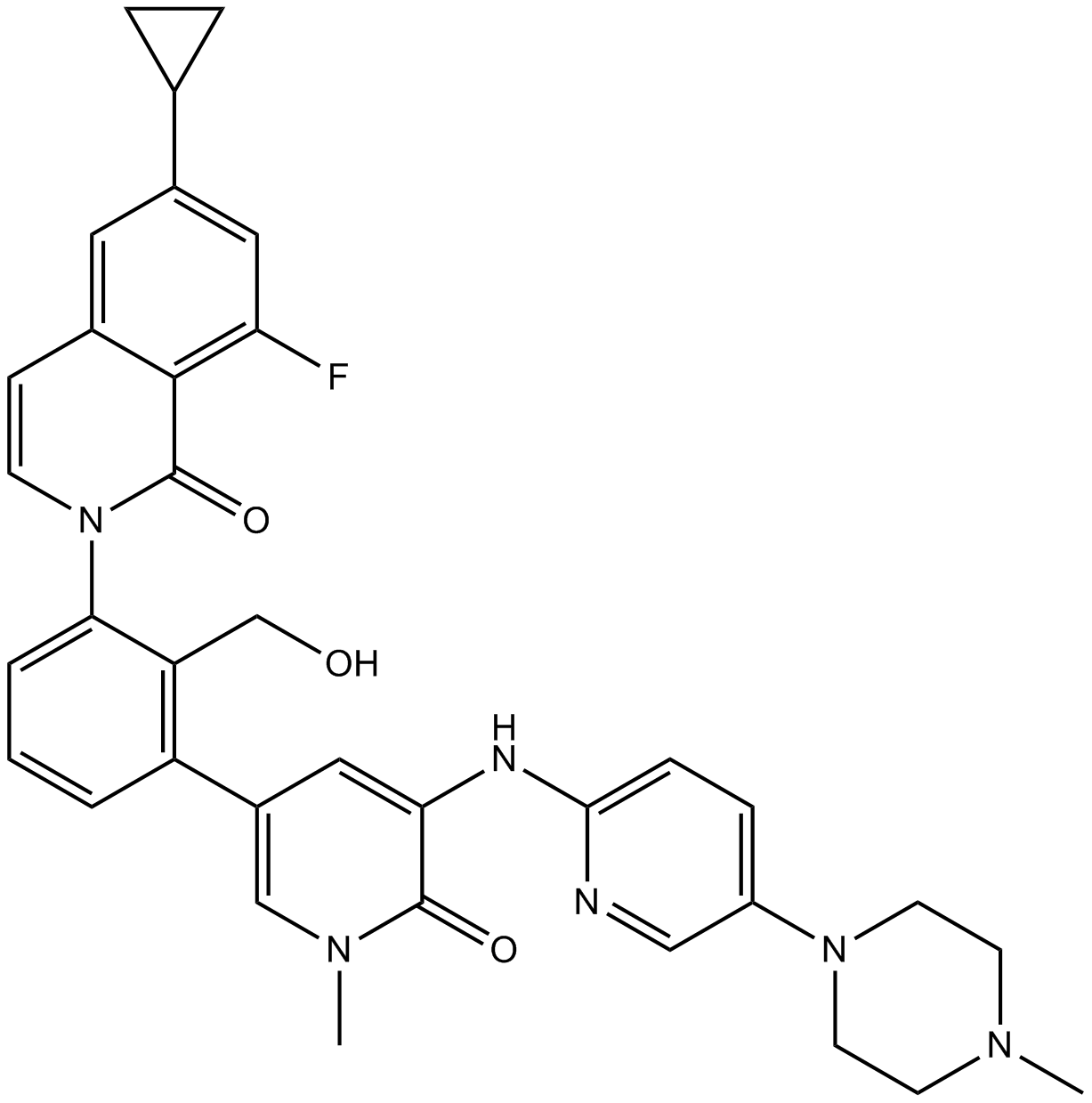
-
GC62397
RO0270608
RO0270608, the active metabolite of R411, is a dual alpha4beta1-alpha4beta7 (α4β1/α4β7) integrin antagonist.

-
GC72461
Rovelizumab
Rovelizumab is a humanized monoclonal leukointegrin antibody.

-
GC14795
RWJ 50271
LFA-1/sICAM interaction inhibitor
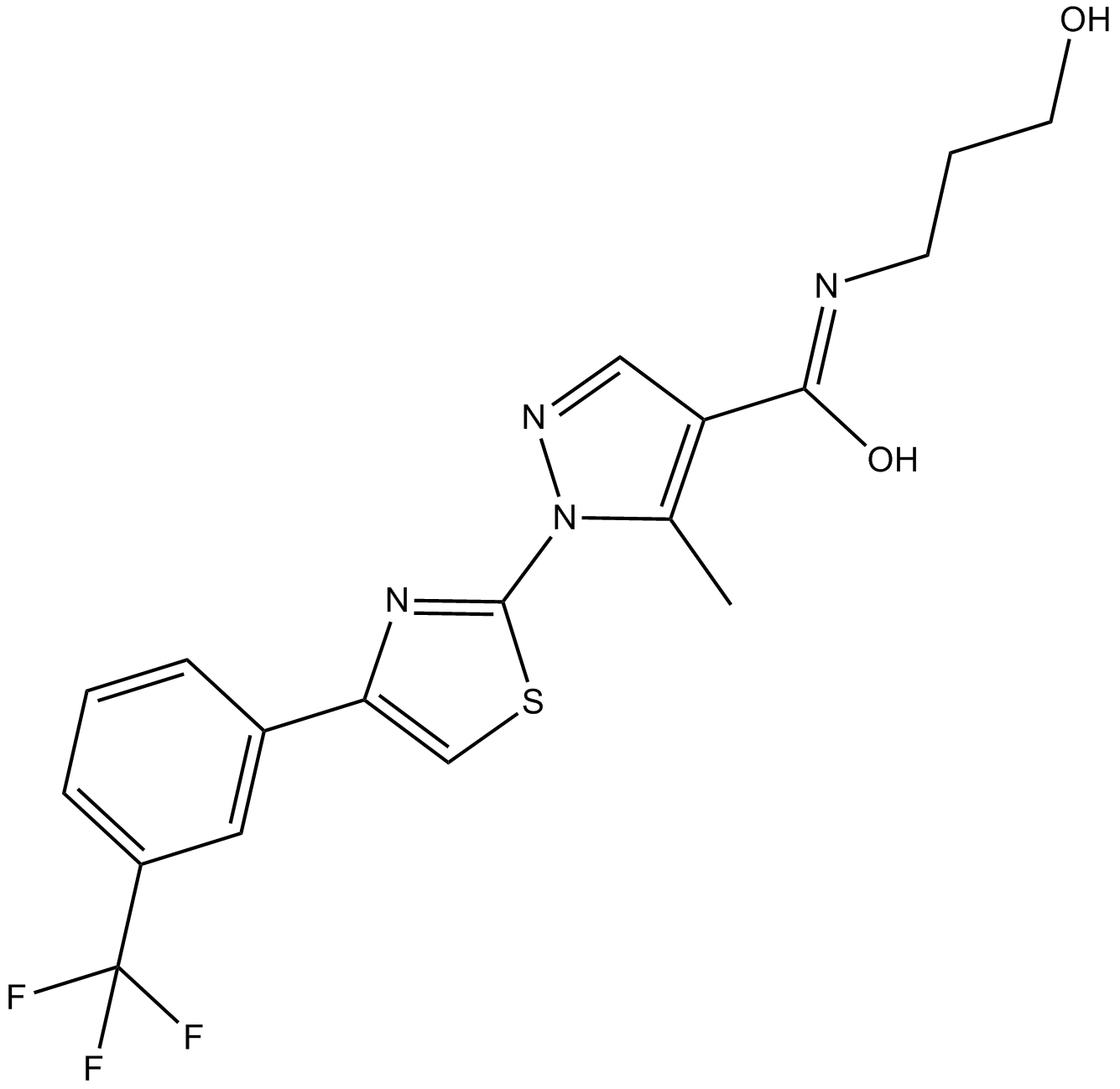
-
GC61777
RWJ-56110 dihydrochloride
RWJ-56110 dihydrochloride is a potent, selective, peptide-mimetic inhibitor of PAR-1 activation and internalization (binding IC50=0.44 uM) and shows no effect on PAR-2, PAR-3, or PAR-4.

-
GC14143
SB273005
An αvβ3 antagonist

-
GC10291
SCH 79797 dihydrochloride
PAR1 receptor antagonist

-
GC63540
SCH79797
SCH79797 is a highly potent, selective nonpeptide protease activated receptor 1 (PAR1) antagonist.
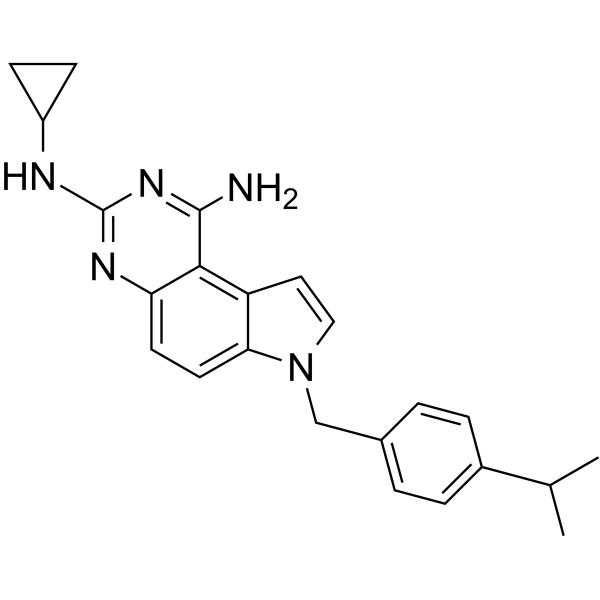
-
GC64571
Sibrafiban
RO 48-3657
Sibrafiban (RO 48-3657) is the orally active, nonpeptide, double-prodrug of Ro 44-3888 and a selective glycoprotein IIb/IIIa receptor antagonist.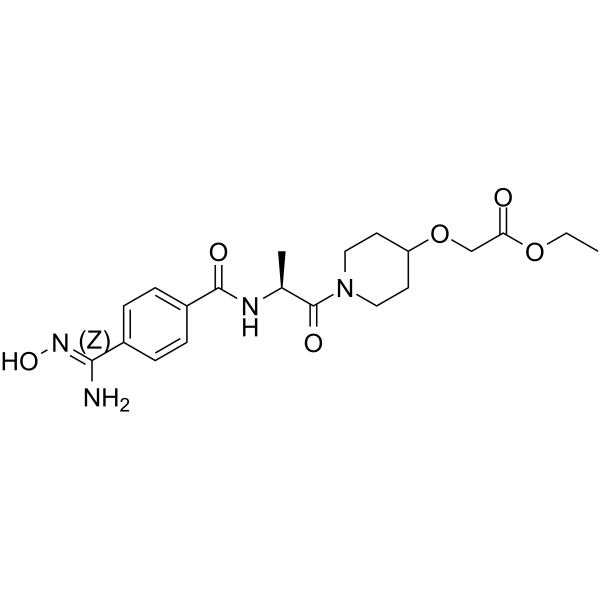
-
GC49713
SIKVAV (acetate)
Hexapeptide-10, Ser-Ile-Lys-Val-Ala-Val
A laminin α1-derived peptide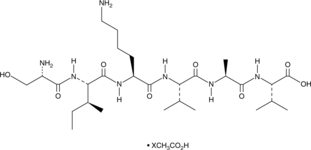
-
GC49358
Sildenafil Chlorosulfonyl
CMP
An intermediate in the synthesis of sildenafil
-
GC48078
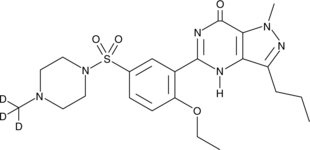
Sildenafil-d3
An internal standard for the quantification of sildenafil
-
GC62614
SJF620 hydrochloride
SJF620 hydrochloride is a PROTAC connected by ligands for Cereblon and Btk with a DC50 of 7.9 nM. SJF620 contains a Lenalidomide analog for recruiting CRBN.

-
GC14540
SLIGKV-NH2
SLIGKV-NH2 (SLIGKV-NH2) is a highly potent protease-activated receptor-2 (PAR2) activating peptide.

-
GC17514
SLIGRL-NH2
Protease-Activated Receptor-2 Activating Peptide
PAR2 activator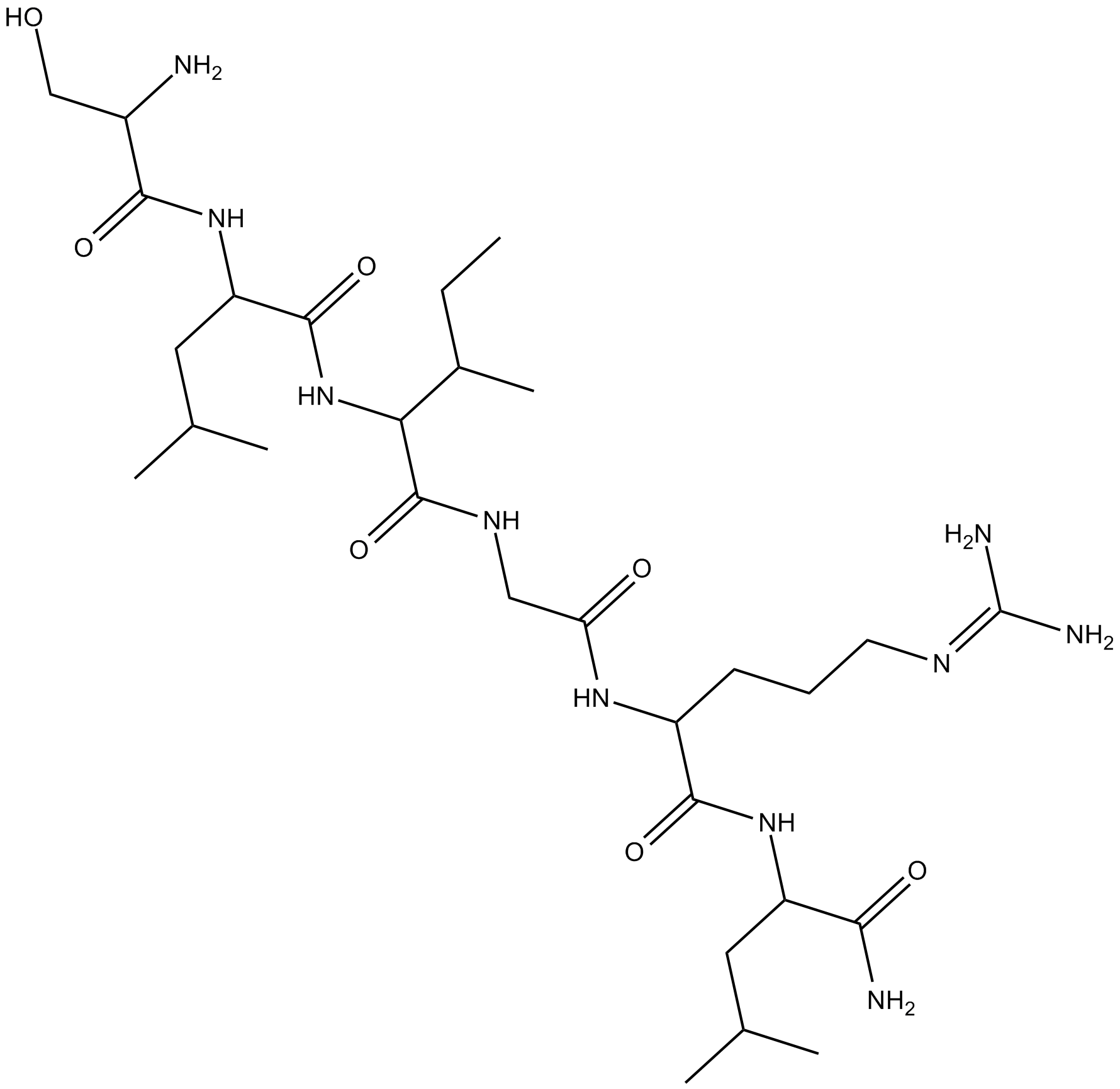
-
GC37672
Spebrutinib besylate
AVL-292 benzenesulfonate; CC-292 besylate
Spebrutinib besylate (AVL-292 benzenesulfonate; CC-292 besylate) is a potent inhibitor of Btk kinase activity (IC50<0.5 nM, Kinact/Ki=7.69×104 M-1s-1s) in biochemical assays.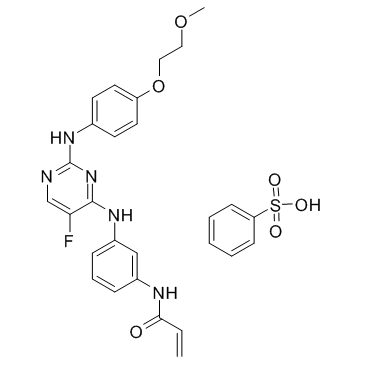
-
GC40963
Spirolaxine
Spirolaxine is a long-chain phthalide produced by the fungus S.
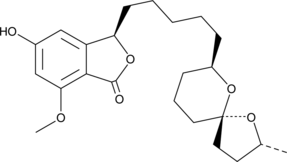
-
GC31993
SR121566A
SR121566A is a novel non-peptide Glycoprotein IIb/IIIa (GP IIb-IIIa) antagonist, which can inhibit ADP-, arachidonic acid- and collagen-induced human platelet aggregation with IC50s of 46±7.5, 56±6 and 42±3 nM, respectively.

-
GC44956
Streptochlorin
SF 2583A
Streptochlorin is a bacterial metabolite originally isolated from Streptomyces sp.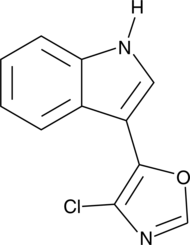
-
GC45688
STX140
2-Methoxyestradiol-bis-sulphamate
An estrogen sulfamate
-
GC44966
Sucrose octasulfate (potassium salt)
SOS, Sucrosofate potassium
Sucrose octasulfate (SOS), a component of the gastrointestinal protectant sucralfate, is an alkaline aluminum-sucrose complex that inhibits peptic hydrolysis and raises gastric pH, protecting esophageal epithelium against acid injury.
-
GC32876
SYP-5
A HIF-1 inhibitor

-
GC64120
TAK-020
TAK-020 is a covalent Btk inhibitor, which becomes the clinical candidate.
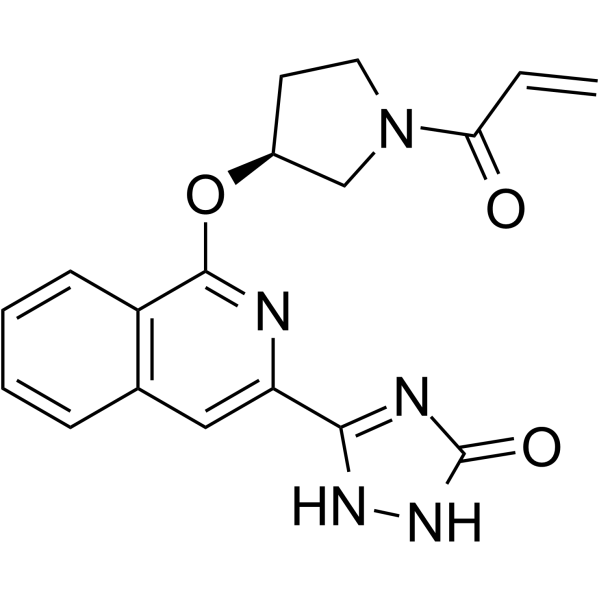
-
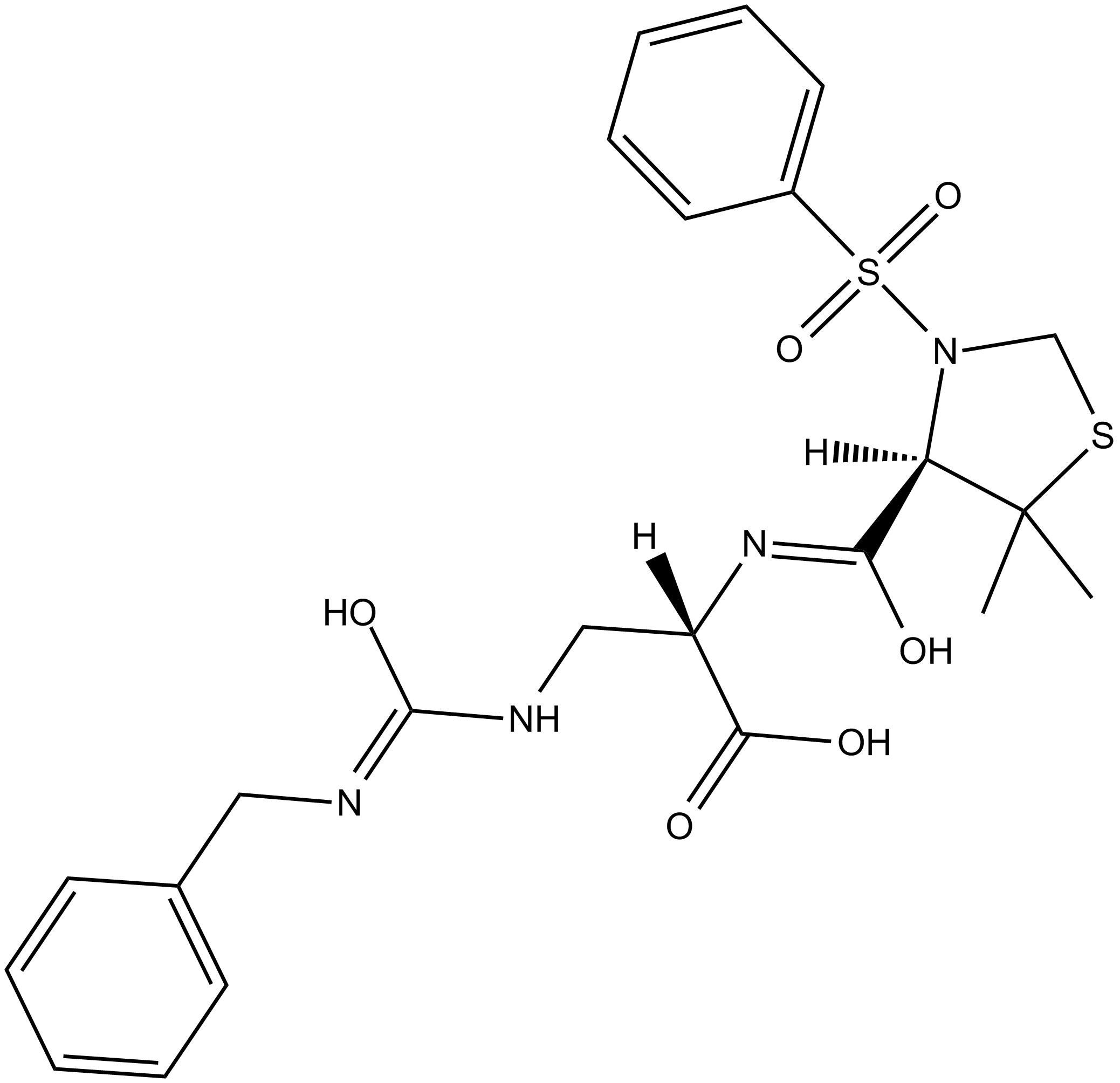
GC12037
TC-I 15
α2β1 integrin inhibitor
-
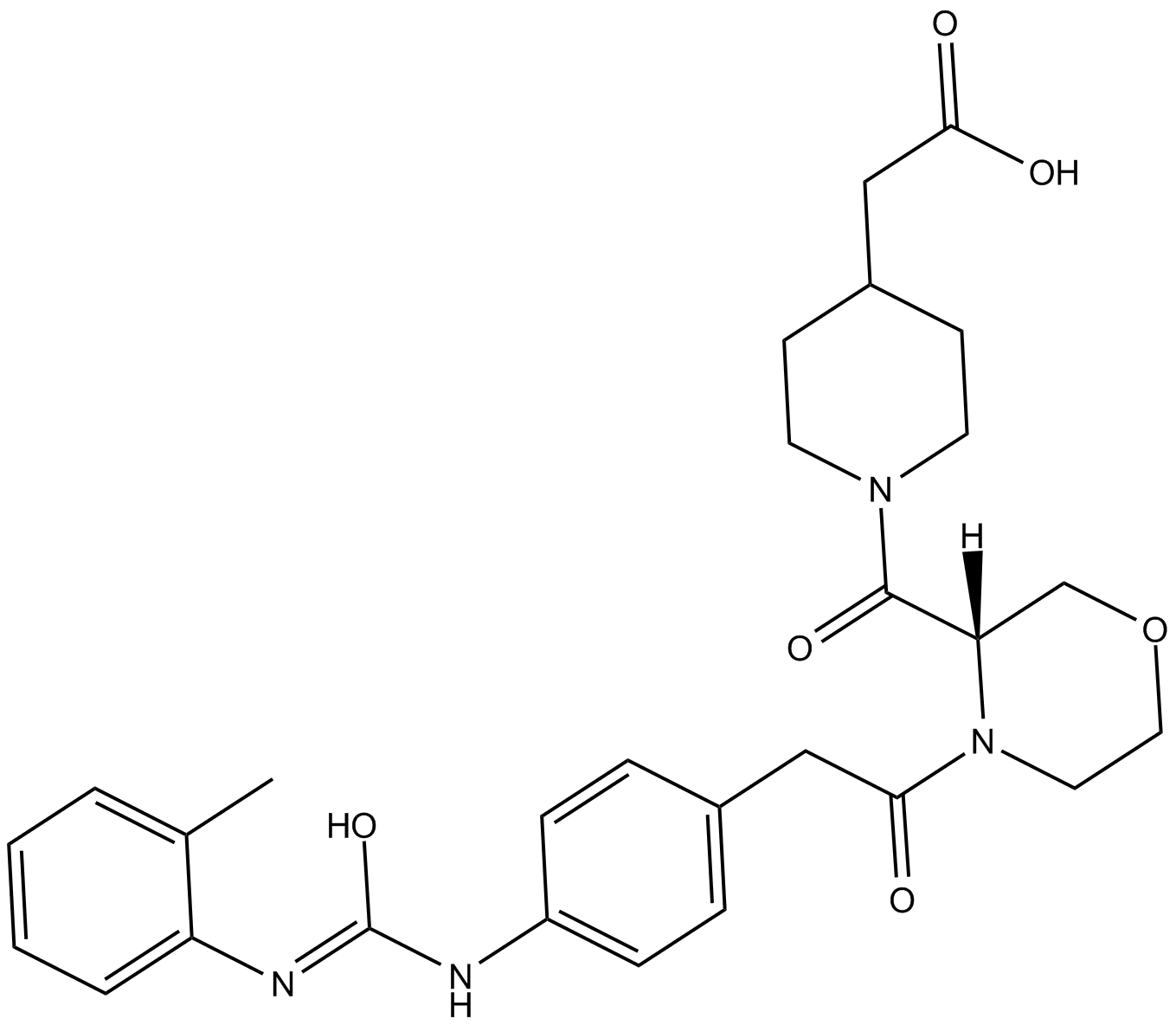
GC18025
TCS 2314
integrin very late antigen-4 (VLA-4; α4β1) antagonist
-
GC61316
tcY-NH2 TFA
(trans-Cinnamoyl)-YPGKF-NH2 TFA
tcY-NH2 ((trans-Cinnamoyl)-YPGKF-NH2) TFA is a potent selective PAR4 antagonist peptide.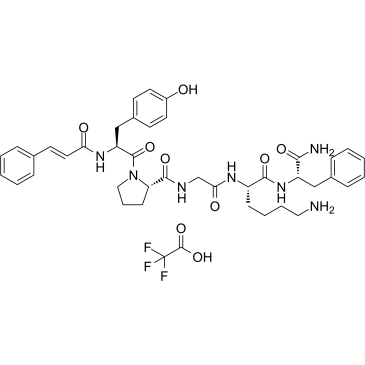
-
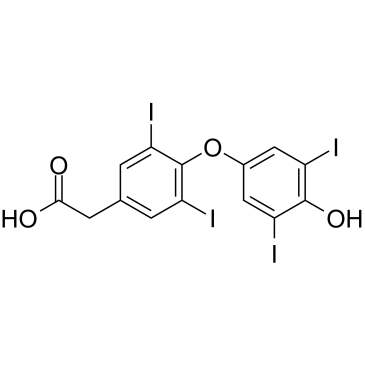
GC61323
Tetrac
Tetrac (Tetraiodothyroacetic acid), a derivative of L-thyroxine (T4), is a thyrointegrin receptor antagonist. Tetrac blocks the actions of T4 and 3,5,3'-triiodo-L-thyronine (T3) at the cell surface receptor for thyroid hormone on integrin αvβ3. Tetra has anti-angiogenic and anti-tumor activities.
-
GC17444
TFLLR-NH2
PAR1 selective agonist
-
GC37770
TFLLR-NH2(TFA)
TFLLR-NH2 (TFA) is a selective PAR1 agonist with an EC50 of 1.9 μM.
-
GP10085
Thrombin Receptor Activator for Peptide 5 (TRAP-5)
H2N-Ser-Phe-Leu-Leu-Arg-OH
-
GC10140
Thrombin Receptor Agonist Peptide
Thrombin Receptor Agonist Peptide is a synthetic thrombin receptor agonist peptide.

-
GC32945
THS-044
THS-044 binding stabilizes the HIF2α PAS-B folded state, for regulating HIF2 activity in endogenous and clinical settings.
-
GC45054
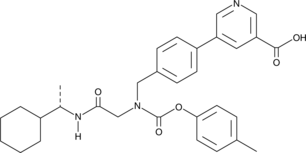
Tie2 Inhibitor 7
Tie2 Inhibitor 7 blocks Tie2 kinase activity with a Ki value of 1.3 μM..
-
GC32228
Tilorone dihydrochloride
NSC 143969
An interferon-inducing antiviral agent
-
GC33821
Tirabrutinib (ONO-4059)
GS-4059, ONO-WG-307, Tirabrutinib
Tirabrutinib (ONO-4059) (ONO-4059) is an orally active Bruton's Tyrosine Kinase (BTK) inhibitor (can cross the blood-brain barrier (BBB)), with an IC50 of 6.8 nM.
-
GC34097
Tirabrutinib hydrochloride (ONO-4059 (hydrochloride))
GS-4059, ONO-WG-307
Tirabrutinib (ONO-4059) hydrochloride is an orally active Bruton's Tyrosine Kinase (BTK) inhibitor (can cross the blood-brain barrier (BBB)), with an IC50 of 6.8 nM.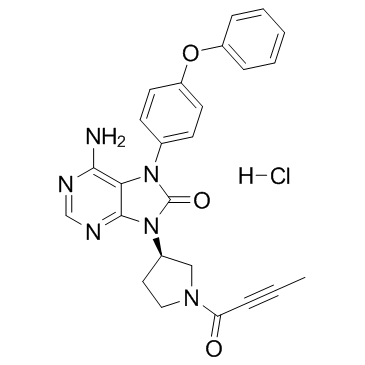
-
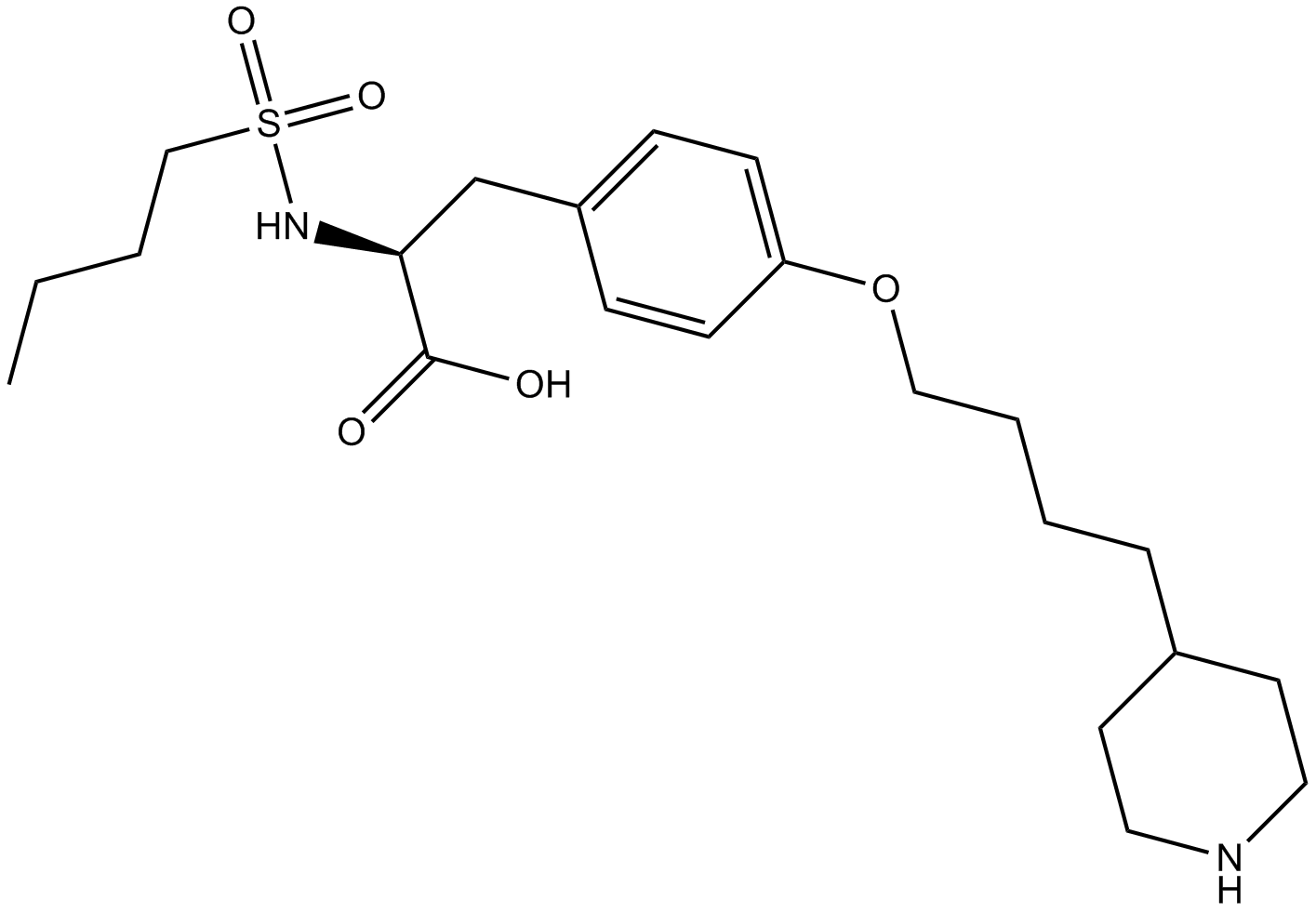
GC13255
Tirofiban
selective platelet GPIIb/IIIa antagonist
-
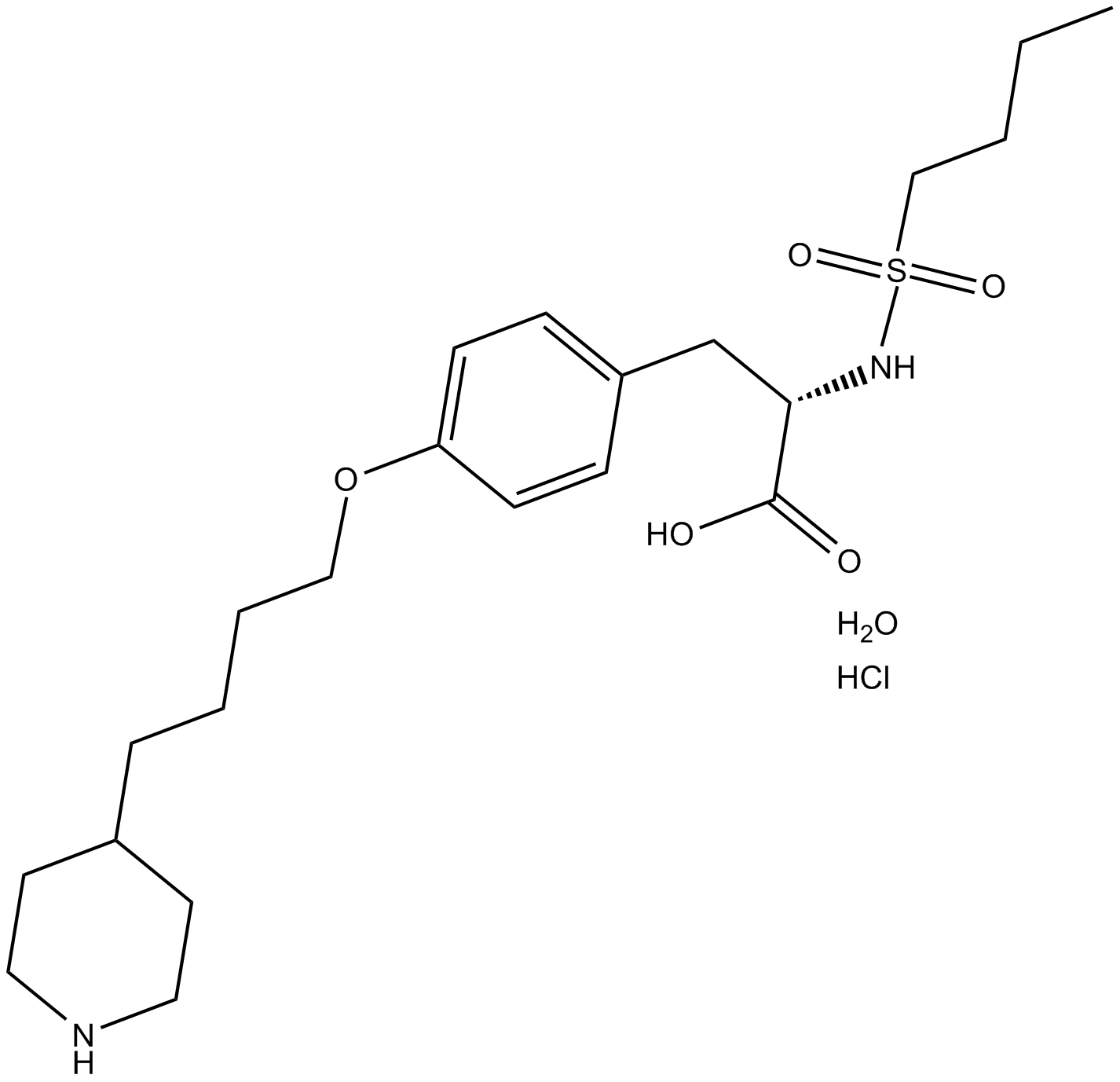
GC11371
Tirofiban hydrochloride monohydrate
A GPIIb/IIIa antagonist
-
GC63229
TL-895
TL-895 is a potent, orally active, ATP-competitive, and highly selective irreversible BTK inhibitor with an IC50 and a Ki of 1.5 nM and 11.9 nM, respectively.

-
GC37801
TM6089
TM6089 is a unique Prolyl Hydroxylase (PHD) inhibitor which stimulates HIF activity without iron chelation and induces angiogenesis and exerts organ protection against ischemia.
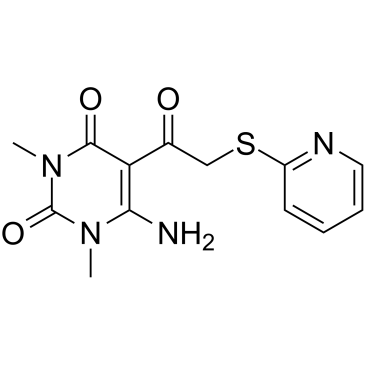
-
GC63232
Tolebrutinib
SAR442168; PRN2246
Tolebrutinib (SAR442168) is a potent, selective, orally active and brain-penetrant inhibitor of Bruton tyrosine kinase (BTK), with IC50s of 0.4 and 0.7 nM in Ramos B cells and in HMC microglia cells, respectively.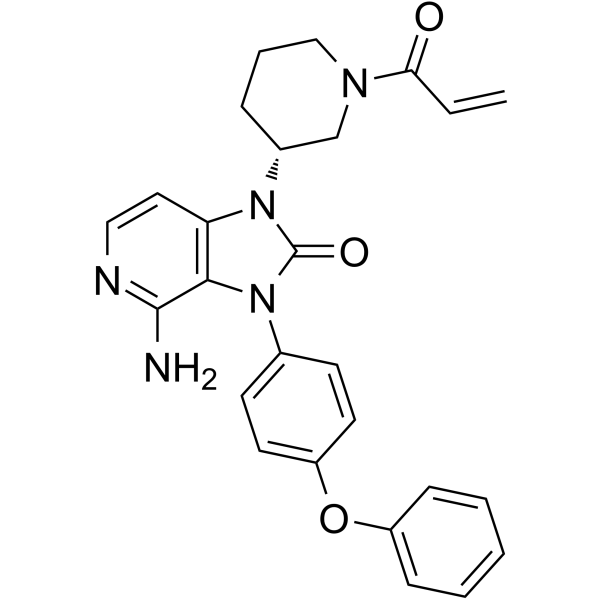
-
GC31498
TP0463518
TP0463518 is a potent hypoxia-inducible factor prolyl hydroxylases (PHDs) inhibitor
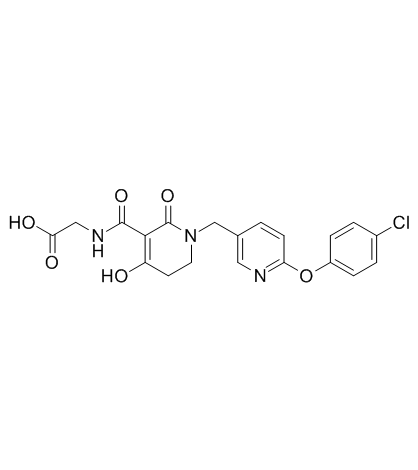
-
GC37818
TR-14035
TR-14035 is a orally active dual α4β7/α4β1 integrin antagonist, with IC50 s of 7 nM and 87 nM for α4β7 and α4β1, respectively.
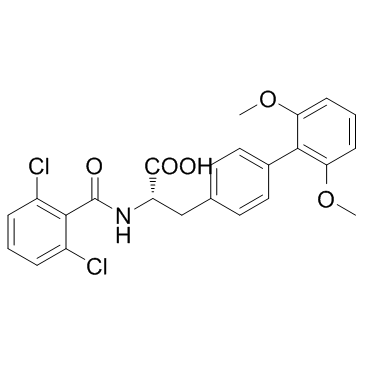
-
GC48195
trans-trismethoxy Resveratrol-d4
(E)-5-2-(4-hydroxyphenyl)ethenyl-1,3-benzene diol-d4, TMS-d4, trans-3,5,4'-Trimethoxystilbene-d4
An internal standard for the quantification of trans-trismethoxy resveratrol
-
GC11662
TRAP-6
PAR-1 agonist peptide; Thrombin Receptor Activator Peptide 6
PAR1 agonist
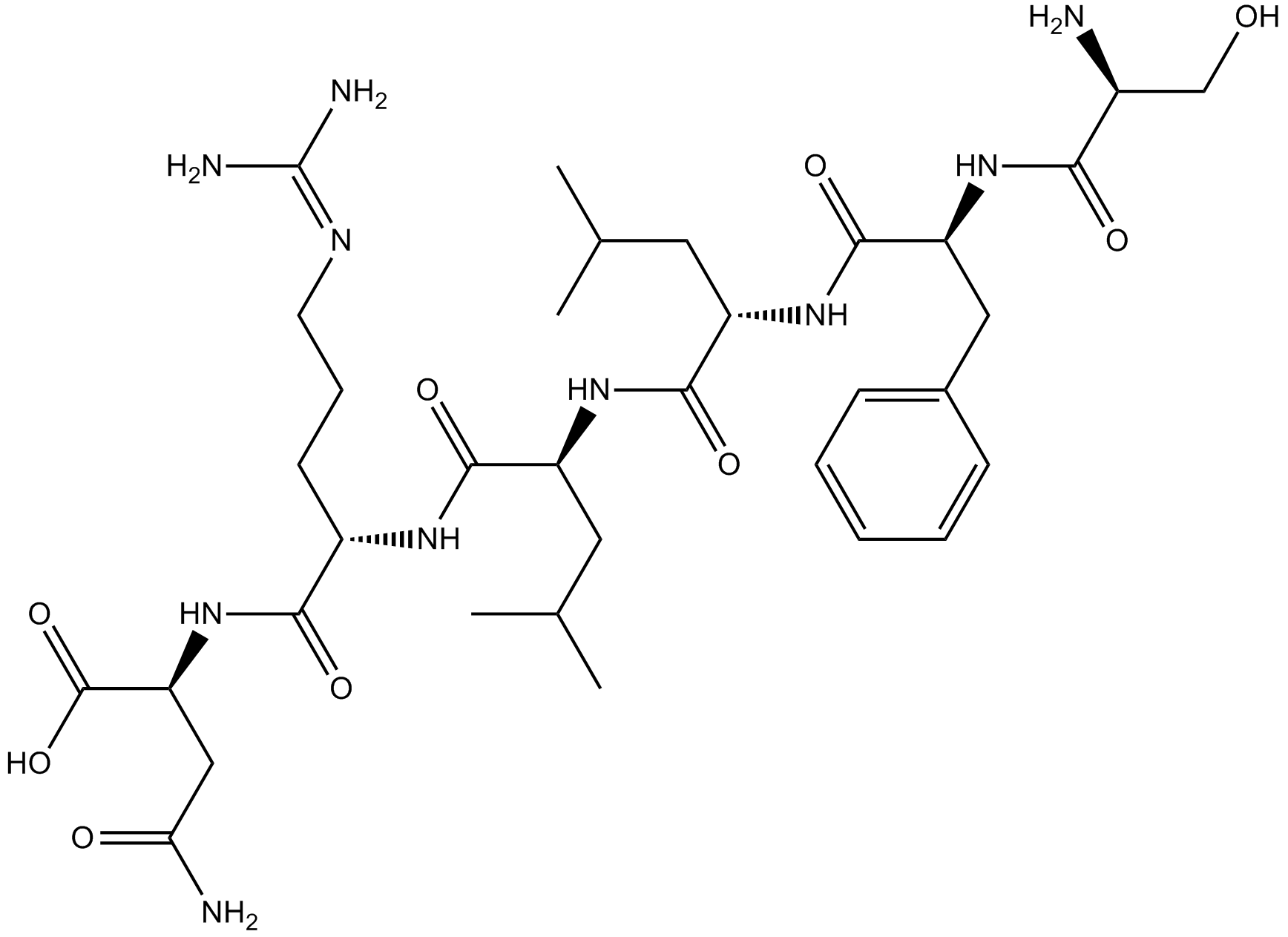
-
GC65529
TRAP-6 amide
TRAP-6 amide is a PAR-1 thrombin receptor agonist peptide.

-
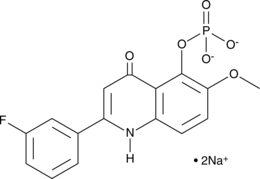
GC48210
TRX-818 (sodium salt)
An anticancer compound
-
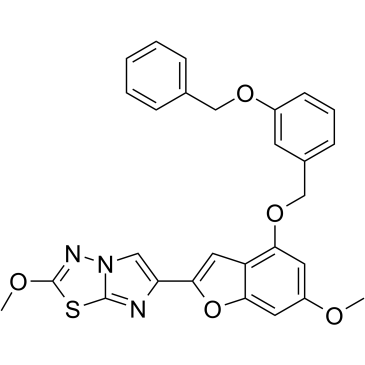
GC37852
UDM-001651
UDM-001651 is a potent, selective, and orally bioavailable protease-activated receptor 4 (PAR4) antagonist (IC50=4 nM; Kd=1.4 nM).
-
GC16811
Vadadustat
AKB6548, PG-1016548
HIF-PH inhibitor
-
GC63490
Valategrast
R-411 free base
Valategrast (R-411 free base) is a potent and orally active integrin α4β1 (VLA-4) and α4β7 dual antagonist.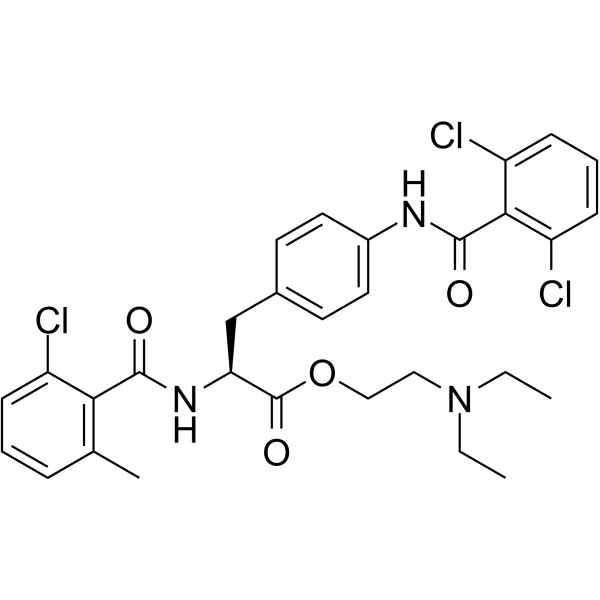
-
GC64212
Valategrast hydrochloride
R-411
Valategrast hydrochloride (R-411) is a potent integrin α4β1 (VLA-4) and α4β7 dual antagonist.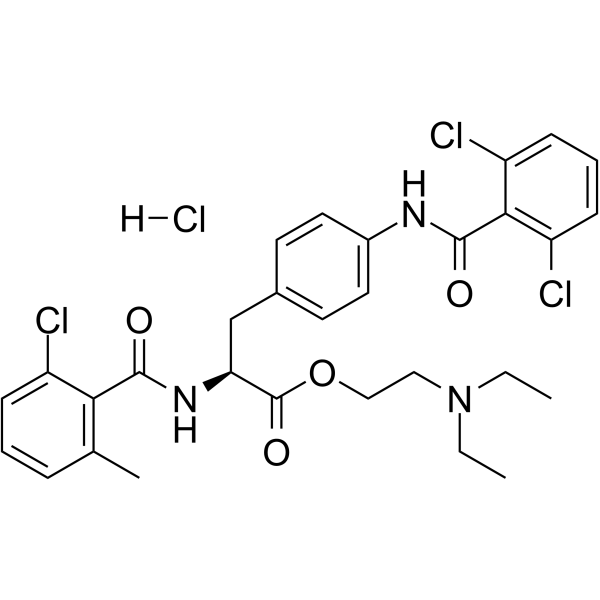
-
GC45769
Vandetanib-d6
ZD6474-d6
An internal standard for the quantification of vandetanib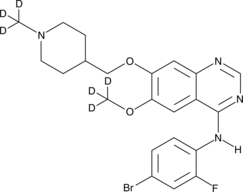
-
GC37889
VCE-004.8
VCE-?004.8
VCE-004.8 (VCE-004.8) is an orally active, specific PPARγ and CB2 receptor dual agonist.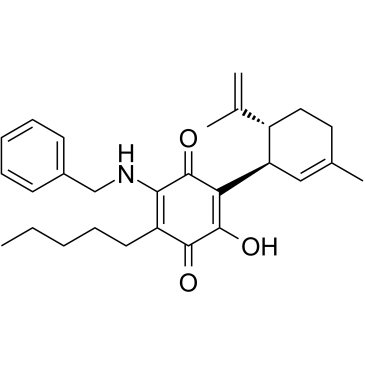
-
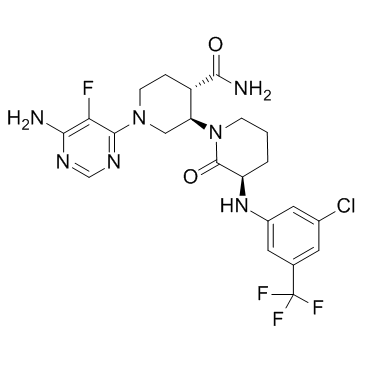
GC34077
Vecabrutinib (SNS-062)
Vecabrutinib (SNS-062) (SNS-062) is a potent, noncovalent BTK and ITK inhibitor, with Kd values of 0.3 nM and 2.2 nM, respectively. Vecabrutinib (SNS-062) shows an IC50 of 24 nM for ITK.
-
GC34211
Vedolizumab
Vedolizumab is a humanized IgG1 monoclonal antibody that targets the α4β7 integrin for the treatment of ulcerative colitis and Crohn's disease.

-
GC11727
VKGILS-NH2
control peptide for SLIGKV-NH2, PAR1 agonist