Apoptosis
As one of the cellular death mechanisms, apoptosis, also known as programmed cell death, can be defined as the process of a proper death of any cell under certain or necessary conditions. Apoptosis is controlled by the interactions between several molecules and responsible for the elimination of unwanted cells from the body.
Many biochemical events and a series of morphological changes occur at the early stage and increasingly continue till the end of apoptosis process. Morphological event cascade including cytoplasmic filament aggregation, nuclear condensation, cellular fragmentation, and plasma membrane blebbing finally results in the formation of apoptotic bodies. Several biochemical changes such as protein modifications/degradations, DNA and chromatin deteriorations, and synthesis of cell surface markers form morphological process during apoptosis.
Apoptosis can be stimulated by two different pathways: (1) intrinsic pathway (or mitochondria pathway) that mainly occurs via release of cytochrome c from the mitochondria and (2) extrinsic pathway when Fas death receptor is activated by a signal coming from the outside of the cell.
Different gene families such as caspases, inhibitor of apoptosis proteins, B cell lymphoma (Bcl)-2 family, tumor necrosis factor (TNF) receptor gene superfamily, or p53 gene are involved and/or collaborate in the process of apoptosis.
Caspase family comprises conserved cysteine aspartic-specific proteases, and members of caspase family are considerably crucial in the regulation of apoptosis. There are 14 different caspases in mammals, and they are basically classified as the initiators including caspase-2, -8, -9, and -10; and the effectors including caspase-3, -6, -7, and -14; and also the cytokine activators including caspase-1, -4, -5, -11, -12, and -13. In vertebrates, caspase-dependent apoptosis occurs through two main interconnected pathways which are intrinsic and extrinsic pathways. The intrinsic or mitochondrial apoptosis pathway can be activated through various cellular stresses that lead to cytochrome c release from the mitochondria and the formation of the apoptosome, comprised of APAF1, cytochrome c, ATP, and caspase-9, resulting in the activation of caspase-9. Active caspase-9 then initiates apoptosis by cleaving and thereby activating executioner caspases. The extrinsic apoptosis pathway is activated through the binding of a ligand to a death receptor, which in turn leads, with the help of the adapter proteins (FADD/TRADD), to recruitment, dimerization, and activation of caspase-8 (or 10). Active caspase-8 (or 10) then either initiates apoptosis directly by cleaving and thereby activating executioner caspase (-3, -6, -7), or activates the intrinsic apoptotic pathway through cleavage of BID to induce efficient cell death. In a heat shock-induced death, caspase-2 induces apoptosis via cleavage of Bid.
Bcl-2 family members are divided into three subfamilies including (i) pro-survival subfamily members (Bcl-2, Bcl-xl, Bcl-W, MCL1, and BFL1/A1), (ii) BH3-only subfamily members (Bad, Bim, Noxa, and Puma9), and (iii) pro-apoptotic mediator subfamily members (Bax and Bak). Following activation of the intrinsic pathway by cellular stress, pro‑apoptotic BCL‑2 homology 3 (BH3)‑only proteins inhibit the anti‑apoptotic proteins Bcl‑2, Bcl-xl, Bcl‑W and MCL1. The subsequent activation and oligomerization of the Bak and Bax result in mitochondrial outer membrane permeabilization (MOMP). This results in the release of cytochrome c and SMAC from the mitochondria. Cytochrome c forms a complex with caspase-9 and APAF1, which leads to the activation of caspase-9. Caspase-9 then activates caspase-3 and caspase-7, resulting in cell death. Inhibition of this process by anti‑apoptotic Bcl‑2 proteins occurs via sequestration of pro‑apoptotic proteins through binding to their BH3 motifs.
One of the most important ways of triggering apoptosis is mediated through death receptors (DRs), which are classified in TNF superfamily. There exist six DRs: DR1 (also called TNFR1); DR2 (also called Fas); DR3, to which VEGI binds; DR4 and DR5, to which TRAIL binds; and DR6, no ligand has yet been identified that binds to DR6. The induction of apoptosis by TNF ligands is initiated by binding to their specific DRs, such as TNFα/TNFR1, FasL /Fas (CD95, DR2), TRAIL (Apo2L)/DR4 (TRAIL-R1) or DR5 (TRAIL-R2). When TNF-α binds to TNFR1, it recruits a protein called TNFR-associated death domain (TRADD) through its death domain (DD). TRADD then recruits a protein called Fas-associated protein with death domain (FADD), which then sequentially activates caspase-8 and caspase-3, and thus apoptosis. Alternatively, TNF-α can activate mitochondria to sequentially release ROS, cytochrome c, and Bax, leading to activation of caspase-9 and caspase-3 and thus apoptosis. Some of the miRNAs can inhibit apoptosis by targeting the death-receptor pathway including miR-21, miR-24, and miR-200c.
p53 has the ability to activate intrinsic and extrinsic pathways of apoptosis by inducing transcription of several proteins like Puma, Bid, Bax, TRAIL-R2, and CD95.
Some inhibitors of apoptosis proteins (IAPs) can inhibit apoptosis indirectly (such as cIAP1/BIRC2, cIAP2/BIRC3) or inhibit caspase directly, such as XIAP/BIRC4 (inhibits caspase-3, -7, -9), and Bruce/BIRC6 (inhibits caspase-3, -6, -7, -8, -9).
Any alterations or abnormalities occurring in apoptotic processes contribute to development of human diseases and malignancies especially cancer.
References:
1.Yağmur Kiraz, Aysun Adan, Melis Kartal Yandim, et al. Major apoptotic mechanisms and genes involved in apoptosis[J]. Tumor Biology, 2016, 37(7):8471.
2.Aggarwal B B, Gupta S C, Kim J H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey.[J]. Blood, 2012, 119(3):651.
3.Ashkenazi A, Fairbrother W J, Leverson J D, et al. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors[J]. Nature Reviews Drug Discovery, 2017.
4.McIlwain D R, Berger T, Mak T W. Caspase functions in cell death and disease[J]. Cold Spring Harbor perspectives in biology, 2013, 5(4): a008656.
5.Ola M S, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis[J]. Molecular and cellular biochemistry, 2011, 351(1-2): 41-58.
What is Apoptosis? The Apoptotic Pathways and the Caspase Cascade
Targets for Apoptosis
- Pyroptosis(31)
- Caspase(54)
- 14.3.3 Proteins(1)
- Apoptosis Inducers(41)
- Bax(8)
- Bcl-2 Family(108)
- Bcl-xL(8)
- c-RET(8)
- IAP(26)
- KEAP1-Nrf2(68)
- MDM2(13)
- p53(112)
- PC-PLC(4)
- PKD(7)
- RasGAP (Ras- P21)(1)
- Survivin(6)
- Thymidylate Synthase(10)
- TNF-α(131)
- Other Apoptosis(884)
- Apoptosis Detection(0)
- Caspase Substrate(0)
- APC(5)
- PD-1/PD-L1 interaction(61)
- ASK1(3)
- PAR4(2)
- RIP kinase(50)
- FKBP(19)
- Cuproptosis(0)
Products for Apoptosis
- Cat.No. Product Name Information
-
GC11543
Cesium chloride

-
GC68857
Cetrelimab
JNJ 63723283; JNJ 3283
Cetrelimab (JNJ 63723283; JNJ 3283) is a humanized IgG4 monoclonal antibody that targets PD-1. Cetrelimab has a Kd of 1.72 nM for binding to PD-1 (HEK293 cells). As such, Cetrelimab blocks the interaction between PD-1 and its ligands, PD-L1 and PD-L2 (with IC50 values of 111.7 ng/mL and 138.6 ng/mL, respectively). Cetrelimab also stimulates peripheral T cells, increases cytokine levels (IFN-gamma, IL-2, TNF-alpha), and inhibits tumor growth in vivo.

-
GC11710
CFM 4
CFM 4 is a potent small molecular antagonist of CARP-1/APC-2 binding. CFM 4 prevents CARP-1 binding with APC-2, causes G2M cell cycle arrest, and induces apoptosis with an IC50 range of 10-15 μM. CFM 4 also suppresses growth of drug-resistant human breast cancer cells.

-
GC35668
CG-200745
CG-200745
CG-200745 (CG-200745) is an orally active, potent pan-HDAC inhibitor which has the hydroxamic acid moiety to bind zinc at the bottom of catalytic pocket. CG-200745 inhibits deacetylation of histone H3 and tubulin. CG-200745 induces the accumulation of p53, promotes p53-dependent transactivation, and enhances the expression of MDM2 and p21 (Waf1/Cip1) proteins. CG-200745 enhances the sensitivity of Gemcitabine-resistant cells to Gemcitabine and 5-Fluorouracil (5-FU; ). CG-200745 induces apoptosis and has anti-tumour effects.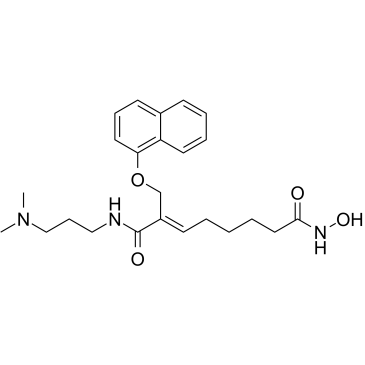
-
GC10666
CGP 57380
MNK1 Inhibitor
MNK1 inhibitor, specific and selective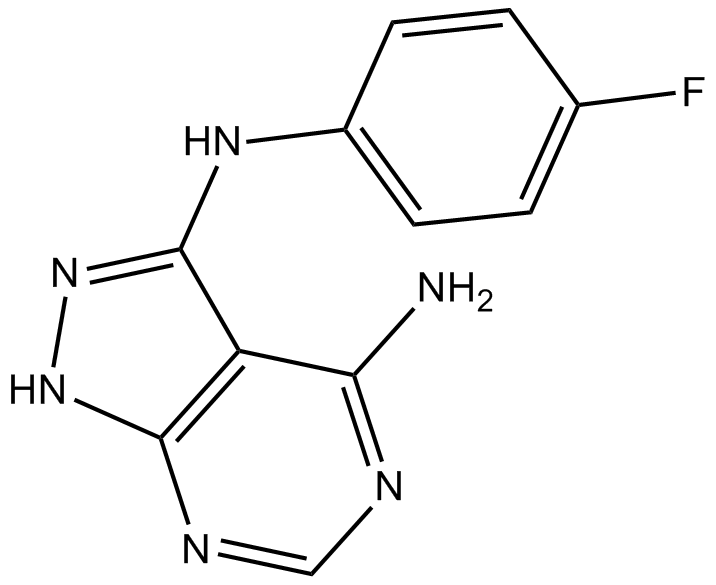
-
GC43234
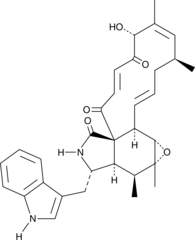
Chaetoglobosin A
Chaetoglobosin A is a mycotoxic cytochalasin that was first isolated from the marine-derived endophytic fungus C.
-
GC18536
Chartreusin
Antibiotic X 465A, Lambdamycin, NSC 5159
Chartreusin is an antibiotic originally isolated from S.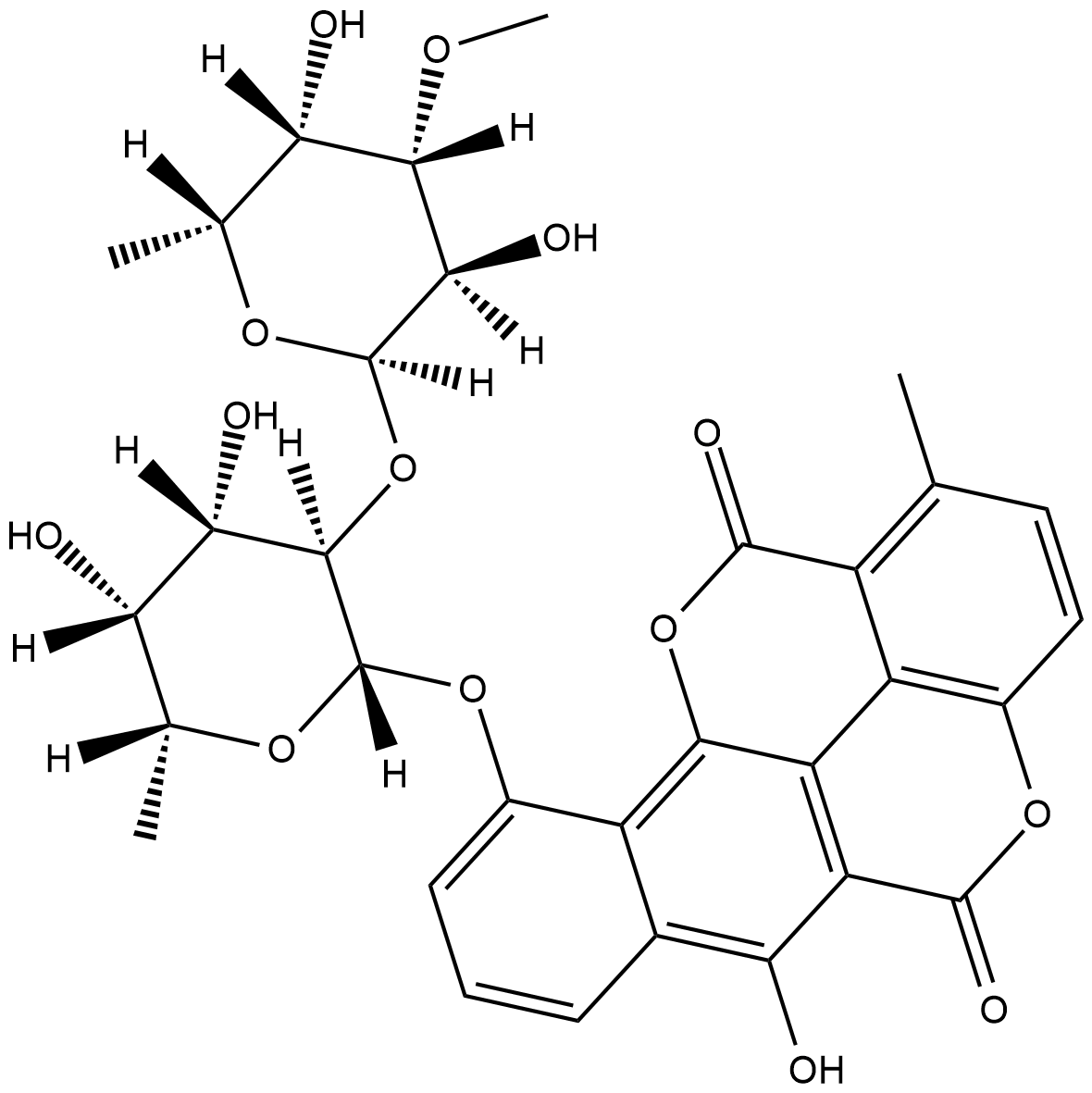
-
GN10463
Chelerythrine

-
GC13065
Chelerythrine Chloride
Broussonpapyrine chloride, NSC 646662
Potent inhibitor of PKC and Bcl-xL
-
GC31886
Chelidonic acid
NSC 3979
A pyran with diverse biological activities
-
GC40878
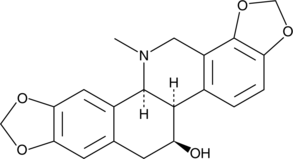
Chelidonine
Chelidonine is a benzophenanthridine alkaloid that has been isolated from C.
-
GC43236
Chevalone B
Chevalone B is a meroterpenoid originally isolated from the fungus E.

-
GC43237
Chevalone C
Chevalone C is a meroterpenoid fungal metabolite originally isolated from E.

-
GC64993
Chicoric acid
L-Chicoric Acid, Dicaffeoyltartaric Acid, NSC 99173
Chicoric acid (Cichoric acid), an orally active dicaffeyltartaric acid, induces reactive oxygen species (ROS) generation.
-
GC15739
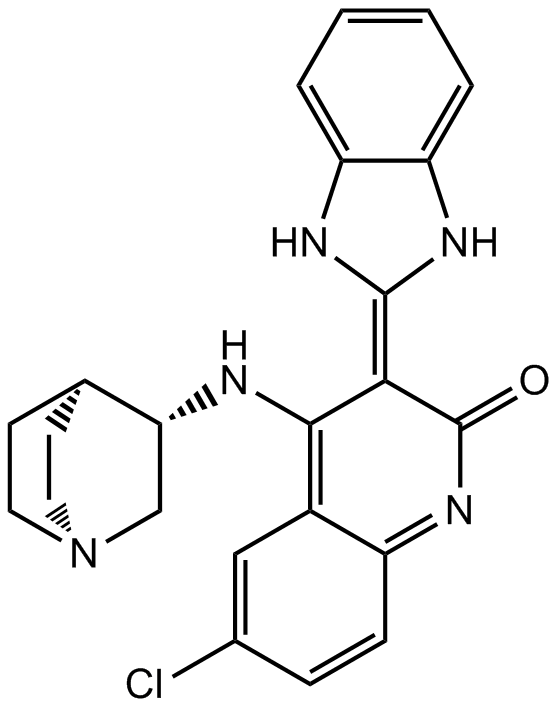
CHIR-124
Chk1 inhibitor,novel and potent
-
GC43239
Chk2 Inhibitor
SC-203885
Chk2 Inhibitor (compound 1) is a potent and selective inhibitor of checkpoint kinase 2 (Chk2), with IC50s of 13.5 nM and 220.4 nM for Chk2 and Chk1, respectively. Chk2 Inhibitor can elicit a strong ataxia telangiectasia mutated (ATM)-dependent Chk2-mediated radioprotection effect.
-
GC45717
Chlamydocin
An HDAC inhibitor

-
GC17969
CHM 1
NSC 656158
An inhibitor of tubulin polymerization
-
GC35682
CHMFL-ABL/KIT-155
CHMFL-ABL-KIT-155
CHMFL-ABL/KIT-155 (CHMFL-ABL-KIT-155; compound 34) is a highly potent and orally active type II ABL/c-KIT dual kinase inhibitor (IC50s of 46 nM and 75 nM, respectively), and it also presents significant inhibitory activities to BLK (IC50=81 nM), CSF1R (IC50=227 nM), DDR1 (IC50=116 nM), DDR2 (IC50=325 nM), LCK (IC50=12 nM) and PDGFRβ (IC50=80 nM) kinases. CHMFL-ABL/KIT-155 (CHMFL-ABL-KIT-155) arrests cell cycle progression and induces apoptosis.
-
GC64028
Chrysosplenol D
Chrysosplenol D is a methoxy flavonoid that induces ERK1/2-mediated apoptosis in triple negative human breast cancer cells.

-
GC13408
CI994 (Tacedinaline)
N-Acetyldinaline, Goe 5549, PD 123654, Tacedinaline
An inhibitor of HDAC1, -2, and -3
-
GC13589
CID 755673
PKD inhibitor

-
GC19436
CID-5721353
CID5721353 is an inhibitor of BCL6 with an IC50 value of 212 μM, which corresponds to a Ki of 147 μM.

-
GC32997
Cinchonine ((8R,9S)-Cinchonine)
Cinchonine ((8R,9S)-Cinchonine) is a natural compound present in Cinchona bark. Cinchonine ((8R,9S)-Cinchonine) activates endoplasmic reticulum stress-induced apoptosis in human liver cancer cells.

-
GC60708
Cinchonine hydrochloride
Cinchonine hydrochloride ((8R,9S)-Cinchonine hydrochloride) is a natural alkaloid present in Cinchona bark, with antimalarial activity. Cinchonine hydrochloride activates endoplasmic reticulum (ER) stress-induced apoptosis in human liver cancer cells.

-
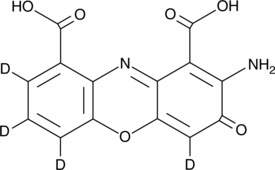
GC52269
Cinnabarinic Acid-d4
An internal standard for the quantification of cinnabarinic acid
-
GC40986
Cinnamamide
NSC 32953
Cinnamamide is an amide form of of trans-cinnamic acid and a metabolite of Streptomyces.
-
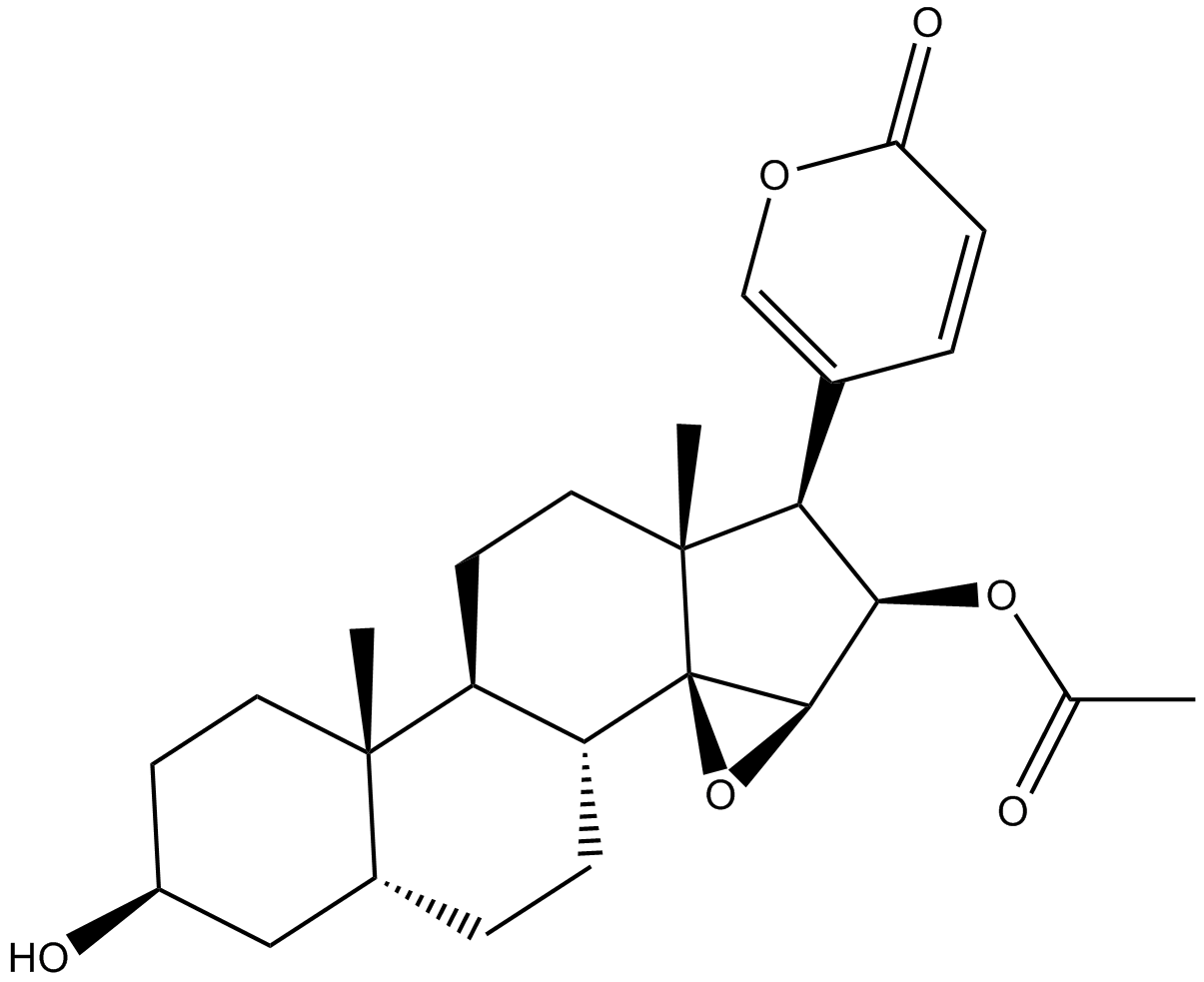
GN10189
Cinobufagin
NSC 90325
-
GC70418
Cipepofol
Cipepofol (Ciprofol), a novel 2,6-disubstituted phenol derivative, is a positive allosteric modulator and direct agonist of the GABAA receptor.

-
GC11908
Cisplatin
CDDP
Cisplatin is one of the best and first metal-based chemotherapeutic drugs, which is used for wide range of solid cancers such as testicular, ovarian, bladder, lung, cervical, head and neck cancer, gastric cancer and some other cancers.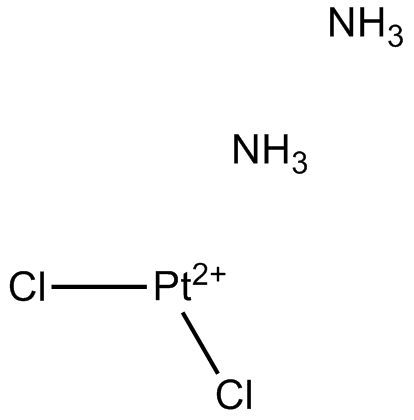
-
GC17491
CITCO
Constitutive androstane receptor (CAR) agonist

-
GC35703
Citicoline
Citicoline (Cytidine diphosphate-choline) is an intermediate in the synthesis of phosphatidylcholine, a component of cell membranes.

-
GC31186
Citicoline sodium salt
Cytidine 5'diphosphocholine, Flussorex, Gerolin, Logan, Neurotron, Sinkron
Citicoline sodium salt salt is an intermediate in the synthesis of phosphatidylcholine which is a component of cell membranes and also exerts neuroprotective effects.
-
GC43273
Citreoindole
Citreoindole is a diketopiperazine metabolite isolated from a hybrid cell fusion of two strains of P.

-
GC41514
Citreoviridin
Citreoviridin is a mycotoxin isolated from several Penicillium species that has been shown to inhibit the mitochondrial ATP synthetase system.

-
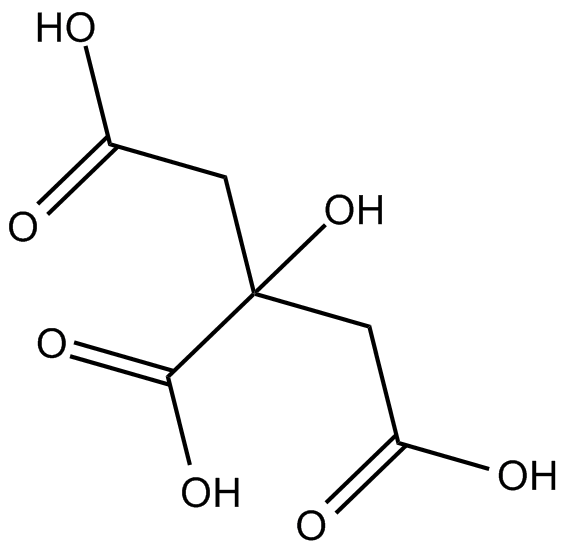
GC14203
Citric acid
Commonly used laboratory reagent
-
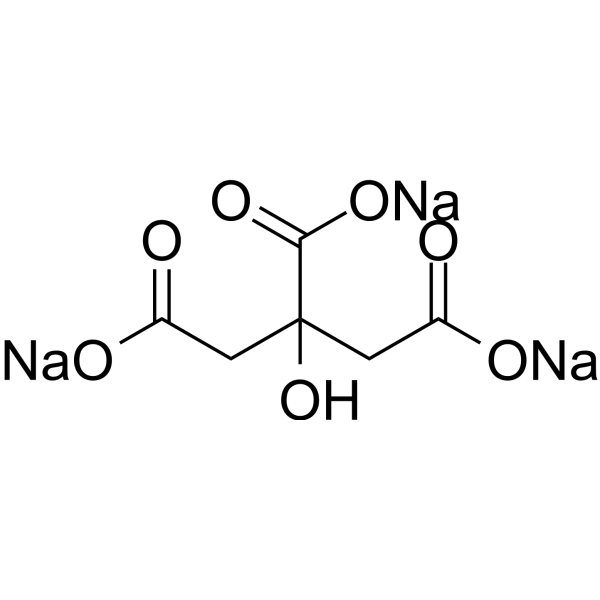
GC68873
Citric acid trisodium
Citric acid trisodium is a natural preservative and food acidifier. It induces apoptosis and cell cycle arrest at the G2/M phase and S phase. Citric acid trisodium causes oxidative damage to the liver by reducing antioxidant enzyme activity. It also causes nephrotoxicity in mice.
-
GC68051
Citric acid-d4

-
GC16661
Citrinin
NSC 186
Citrinin is a mycotoxin with multiple biological activities produced by several fungal strains of the genera Penicillium, Aspergillus, and Monascus.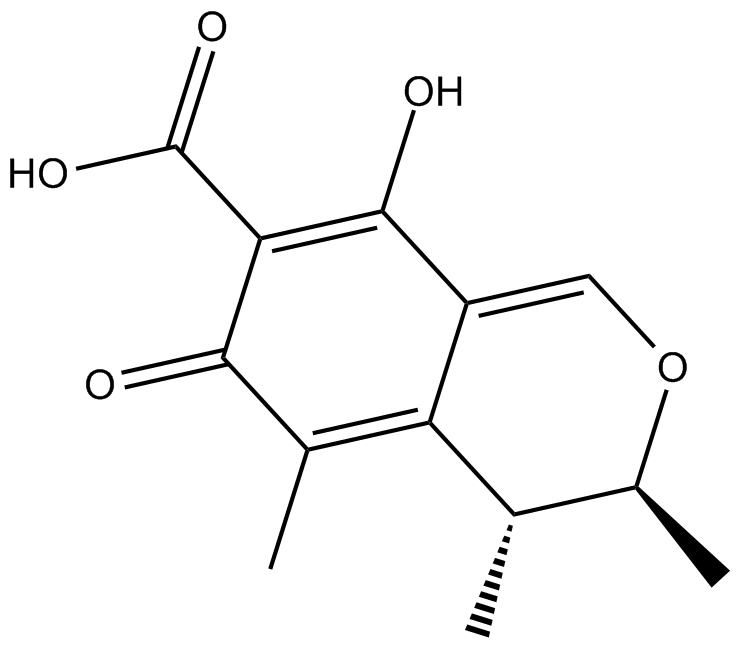
-
GC43274
Citromycetin
NSC 53584
Citromycetin is a fungal metabolite originally isolated from P.
-
GC63393
Citronellyl acetate
Citronellyl acetate is a monoterpene product of the secondary metabolism of plants, with antinociceptive activity.

-
GC52367
Citrullinated Vimentin (G146R) (R144 + R146) (139-159)-biotin Peptide
Biotin-GQGKS(Cit)L(Cit)DLYEEEMRELRRQ, Biotin-GQGKSXLXDLYEEEMRELRRQ (X=Citrulline), Citrullinated VIM (G146R) (R144 + R146)-biotin
A biotinylated and citrullinated mutant vimentin peptide
-
GC52370
Citrullinated Vimentin (R144) (139-159)-biotin Peptide
Biotin-GQGKS(Cit)LGDLYEEEMRELRRQ, Biotin-GQGKSXLGDLYEEEMRELRRQ (X=Citrulline), Citrullinated VIM (R144)-biotin
A biotinylated and citrullinated vimentin peptide
-
GN10219
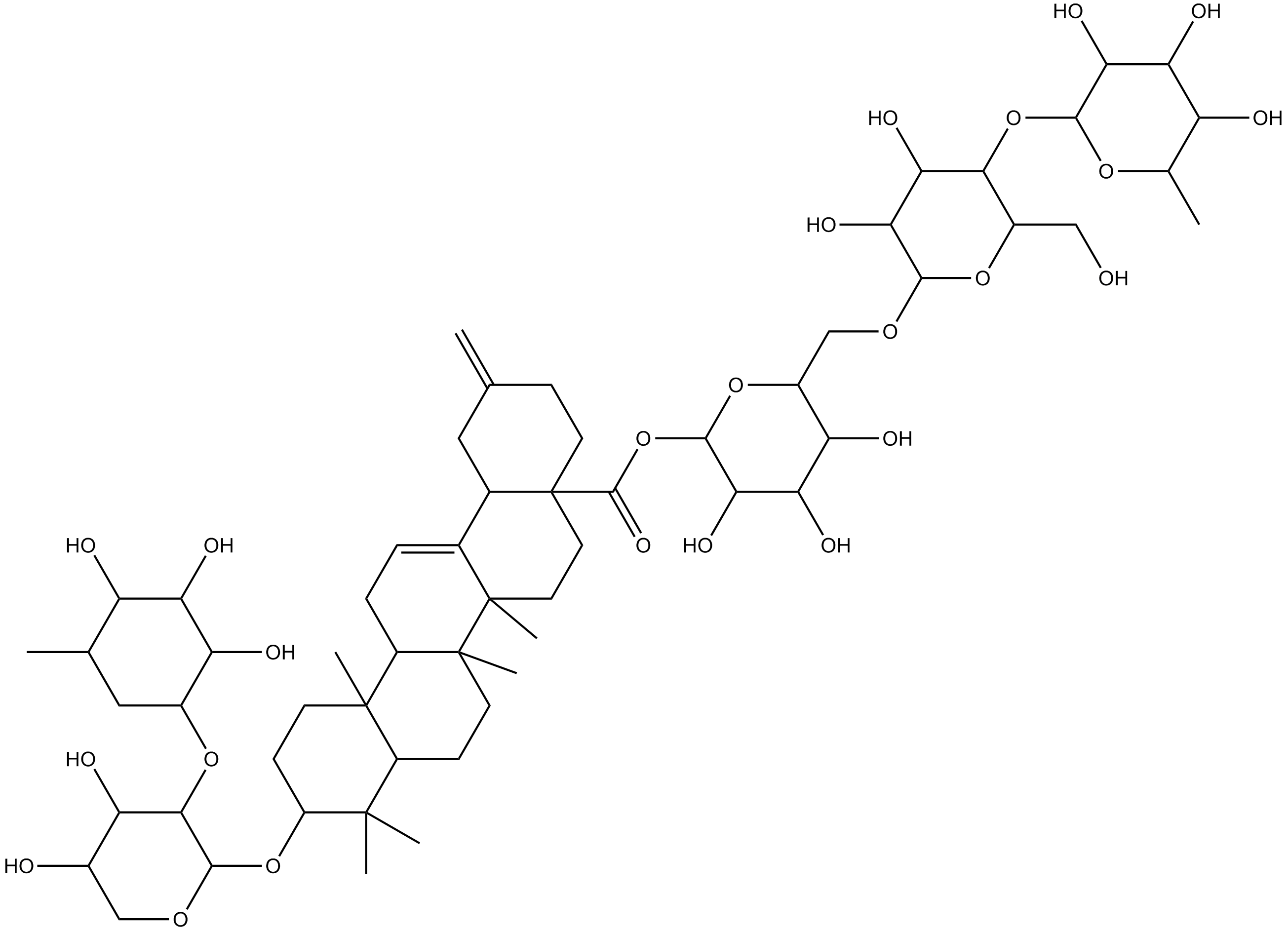
Ciwujianoside-B
-
GC64649
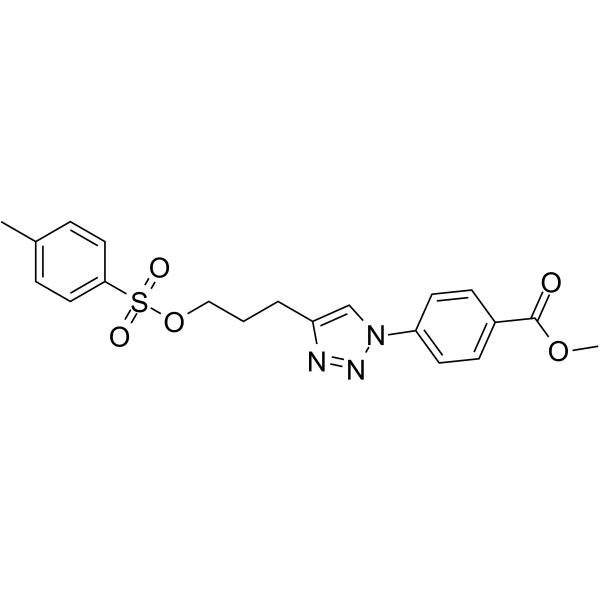
Cjoc42
Cjoc42 is a compound capable of binding to gankyrin. Cjoc42 inhibits gankyrin activity in a dose-dependent manner. Cjoc42 prevents the decrease in p53 protein levels normally associated with high amounts of gankyrin. Cjoc42 restores p53-dependent transcription and sensitivity to DNA damage.
-
GC39485
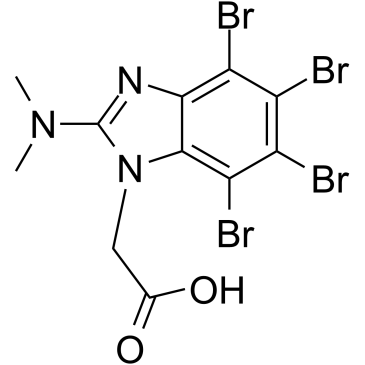
CK2/ERK8-IN-1
A dual inhibitor of CK2 and ERK8
-
GC49556
Cl-Necrostatin-1
7-Cl-Nec-1, 7-Cl-Necrostatin-1, Nec-1f
A RIPK1 inhibitor
-
GC47098
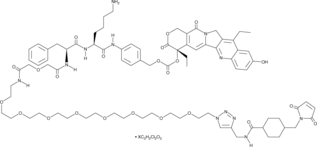
CL2-SN-38 (dichloroacetic acid salt)
An antibody-drug conjugate containing SN-38
-
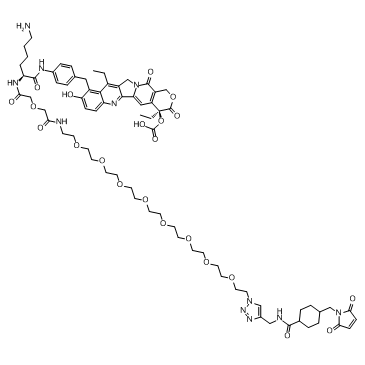
GC60714
CL2A-SN-38
An antibody-drug conjugate containing SN-38
-
GC52469
CL2A-SN-38 (dichloroacetic acid salt)
An antibody-drug conjugate containing SN-38

-
GC10509
Cladribine
2-Chlorodeoxyadenosine, Jk 6251, NSC 105014, RWJ 26251
Apoptosis inducer in CLL cells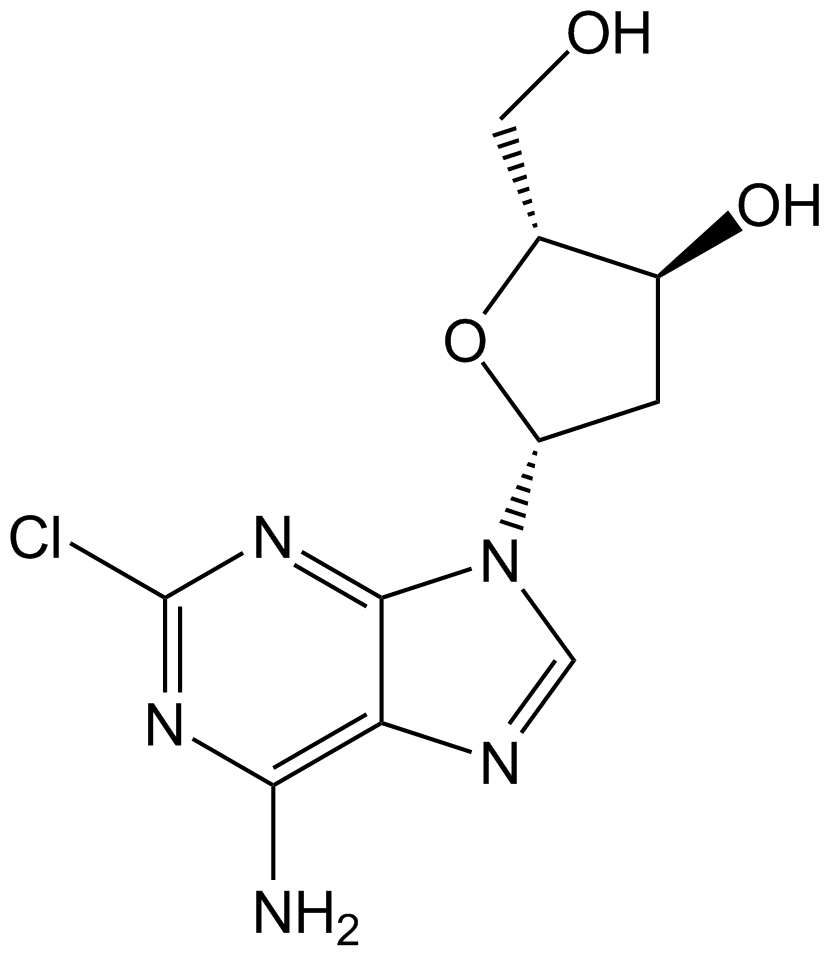
-
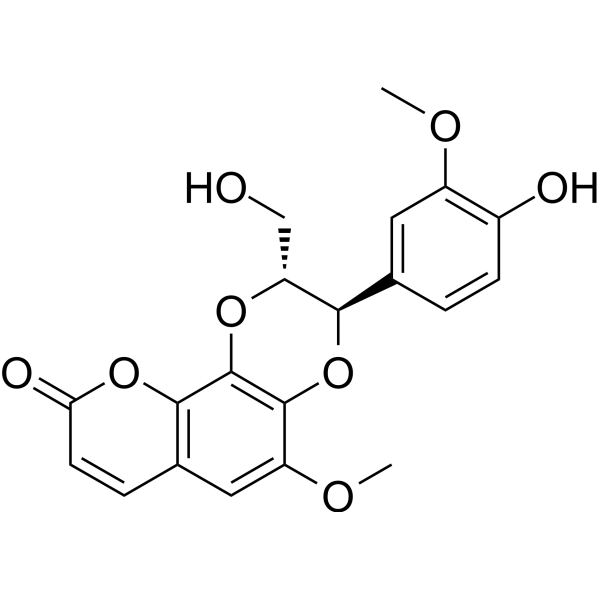
GC68882
Cleomiscosin A
Cleomiscosin A is a coumarin lignan produced by Macaranga adenantha. It has activity in inducing TNF-alpha secretion from mouse peritoneal macrophages.
-
GC60111
Clitocine
Clitocine, an adenosine nucleoside analog isolated from mushroom, is a potent and efficacious readthrough agent. Clitocine acts as a suppressor of nonsense mutations and can induce the production of p53 protein in cells harboring p53 nonsense-mutated alleles. Clitocine can induce apoptosis in multidrug-resistant human cancer cells by targeting Mcl-1. Anticancer activity.

-
GC15219
Clofarabine
Clolar, Evoltra
Antimetabolite,inhibit DNA polymerase and ribonucleotide reductase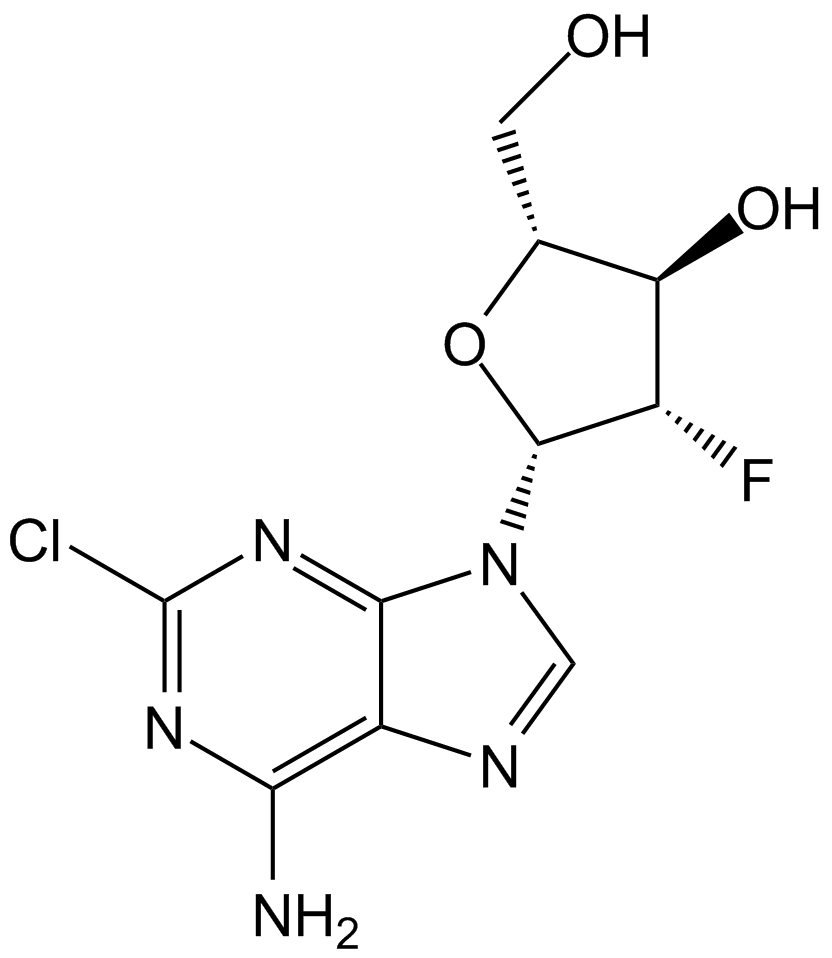
-
GC10813
Clofibric Acid
NSC 1149
PPAR agonist
-
GC32587
Clofilium tosylate
Clofilium tosylate, a potassium channel blocker, induces apoptosis of human promyelocytic leukemia (HL-60) cells via Bcl-2-insensitive activation of caspase-3.

-
GC47105
Clonostachydiol
A fungal metabolite with anticancer and anthelmintic activities
-
GC12367
CM-272
CM-272 is a first-in-class reversible dual inhibitor against G9a and DNMTs with IC50 values of 8 nM and 382 nM, respectively [1].

-
GC62347
CMC2.24
TRB-N0224
CMC2.24 (TRB-N0224), an orally active tricarbonylmethane agent, is effective against pancreatic tumor in mice by inhibiting Ras activation and its downstream effector ERK1/2 pathway.
-
GC61567
CMLD-2
CMLD-2, an inhibitor of HuR-ARE interaction, competitively binds HuR protein disrupting its interaction with adenine-uridine rich elements (ARE)-containing mRNAs (Ki=350 nM). CMLD-2 induces apoptosis exhibits antitumor activity in different cancer cells as colon, pancreatic, thyroid and lung cancer cell lines. Hu antigen R (HuR) is an RNA binding protein, can regulate target mRNAs stability and translation.
-
GC49096
Cobaltic Protoporphyrin IX (chloride)
An inducer of HO-1 activity

-
GC10033
Cobimetinib
GDC-0973, RG-7420, XL518
A potent, orally available MEK1 inhibitor
-
GC43297
Coenzyme Q2
CoQ2, Ubiquinone-2, Ubiquinone Q2
Coenzyme Q10 is a component of the electron transport chain and participates in aerobic cellular respiration, generating energy in the form of ATP.
-
GC62192
COG1410
COG1410 is an apolipoprotein E-derived peptide.
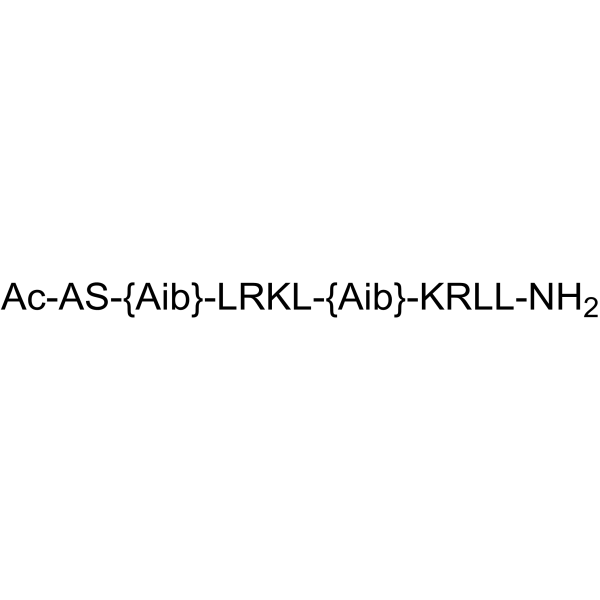
-
GC40664
Colcemid
Demecolcine, NSC 3096
Colcemid is a cytoskeletal inhibitor that induces mitotic arrest in the G2/M phase or meiotic arrest in the vesicle rupture (GVBD) phase in mammalian cells or oocytes, respectively.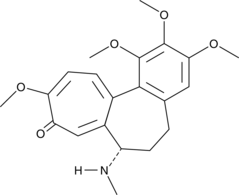
-
GN10123
Columbianadin
Columbianetin
-
GC49454
Complex 3
A fluorescent copper complex with anticancer activity

-
GC18572
Concanavalin A
CConcanavalin A belongs to the concanamycins, a family of macrolide antibiotics isolated from Streptomyces diastatochromogenes that are highly active and selective inhibitors of the vacuolar proton-ATPase (v-[H+]ATPase).

-
GC18832
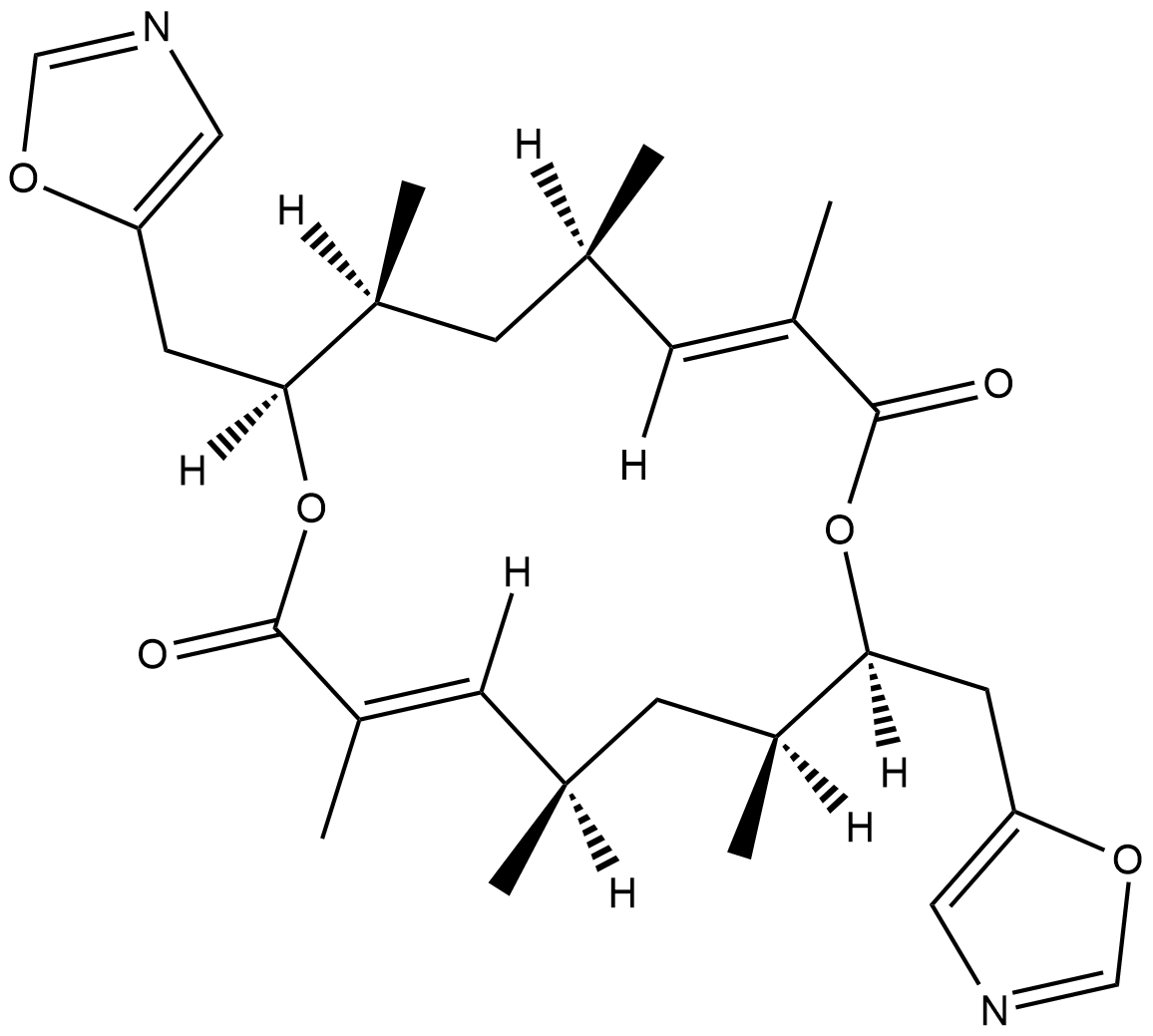
Conglobatin
Conglobatin is a dimeric macrolide dilactone originally isolated from S.
-
GC48483
Conglobatin B
A bacterial metabolite

-
GC48497
Conglobatin C1
A bacterial metabolite
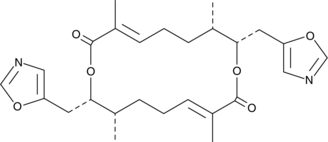
-
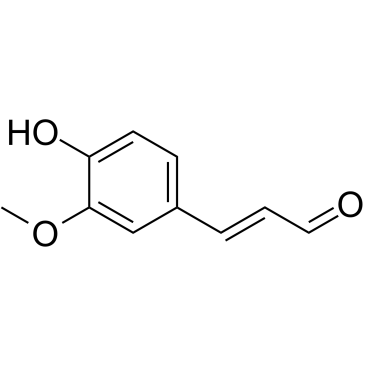
GC38376
Coniferaldehyde
Coniferaldehyde (Ferulaldehyde) is an effective inducer of heme oxygenase-1 (HO-1).
-
GC63379
Conophylline
Conophylline is a vinca alkaloid extracted from leaves of a tropical plant Ervatamia microphylla.

-
GC16772
Cortisone acetate
Cortisone 21-acetate, NSC 49420
Glucocorticoid receptor agonist
-
GC16116
Costunolide
Costus lactone, Melampolide, NSC 106404
A natural sesquiterpene lactone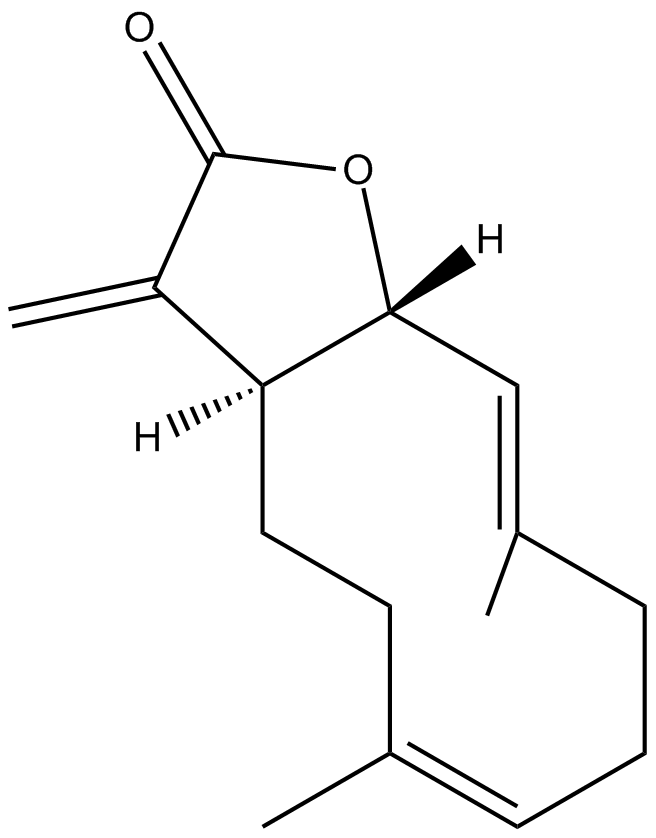
-
GC15225
COTI-2
activates mutant forms of p53

-
GC15840
CP 31398 dihydrochloride
A p53 stabilizing agent

-
GC13091
CP-724714
HER2 inhibitor,potent and selective

-
GC14500
CPI-1189
necrosis factor (TNF) alpha inhibitor
-
GC14699
CPI-203
BET bromodomain inhibitor
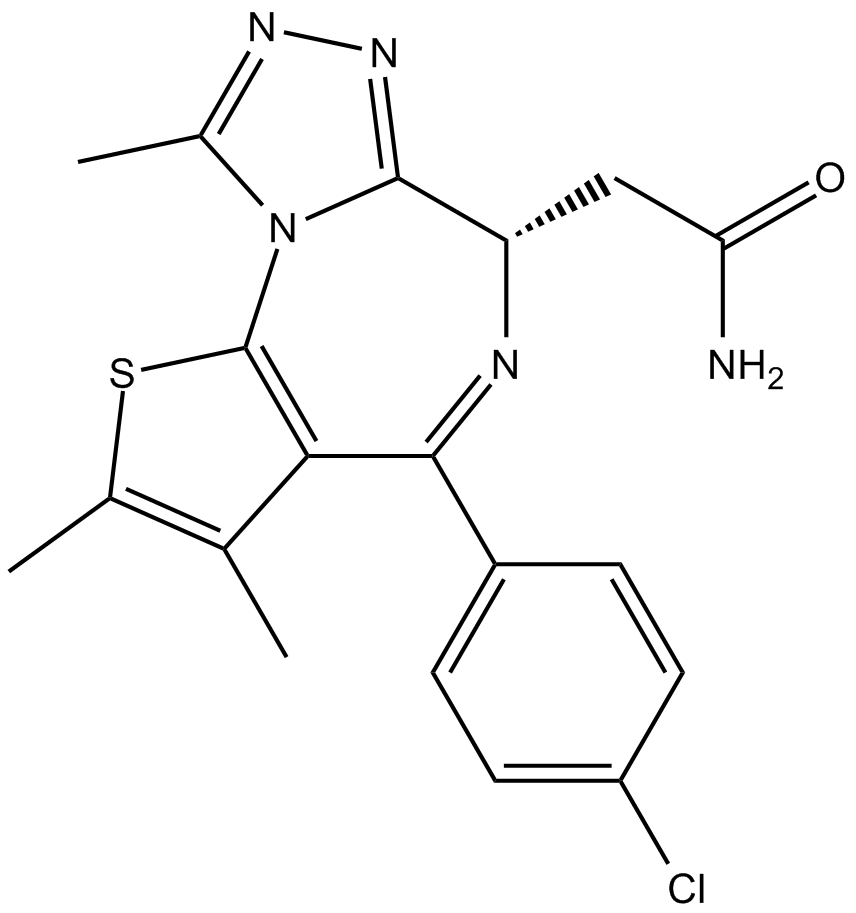
-
GC10021
CPI-360
EZH2 inhibitor

-
GC14921
CPI-613
An inhibitor of α-ketoglutarate dehydrogenase

-
GC39365
CPTH2
CPTH2 is a potent histone acetyltransferase (HAT) inhibitor. CPTH2 selectively inhibits the acetylation of histone H3 by Gcn5. CPTH2 induces apoptosis and decreases the invasiveness of a clear cell renal carcinoma (ccRCC) cell line through the inhibition of acetyltransferase p300 (KAT3B).

-
GC35747
Crebanine
(–)-Crebanine
Crebanine, an alkaloid from Stephania venosa, induces G1 arrest and apoptosis in human cancer cells.
-
GC34543
cRIPGBM
cRIPGBM, a proapoptotic derivative of RIPGBM, a cell type-selective inducer of apoptosis in GBM cancer stem cells (CSCs) by binding to receptor-interacting protein kinase 2 (RIPK2), with an EC50 of 68 nM in GBM-1 cells.
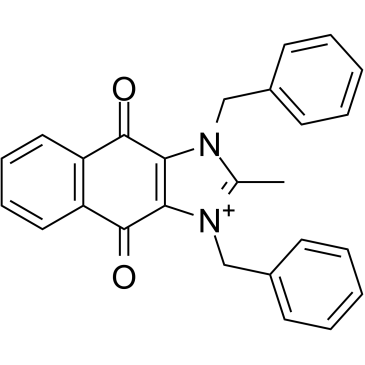
-
GC68903
cRIPGBM chloride
cRIPGBM chloride is an orally effective pro-apoptotic derivative that can be produced from cancer stem cells (CSC) of glioblastoma multiforme (GBM). cRIPGBM chloride induces caspase 1-dependent cell apoptosis by targeting receptor-interacting protein kinase 2 (RIPK2) through the formation of RIPK2/caspase 1 complex and inhibition of RIPK2/TAK1 (pro-survival complex) formation. In animal models, cRIPGBM chloride exhibits strong in vivo anti-tumor activity.
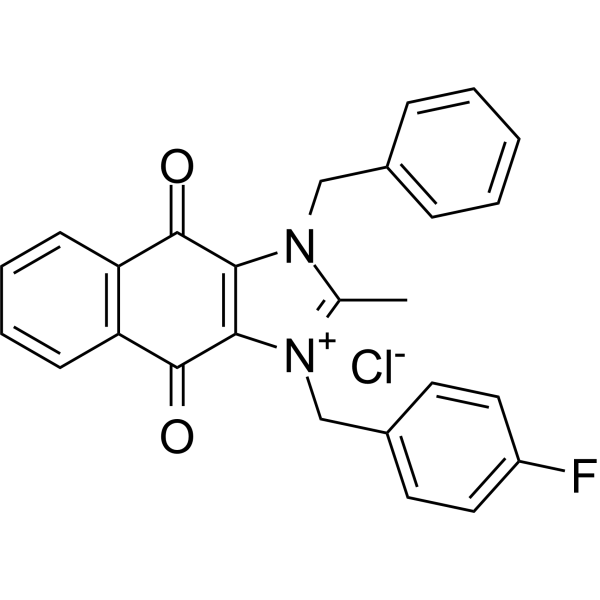
-
GC13838
CRT 0066101
PKD inhibitor
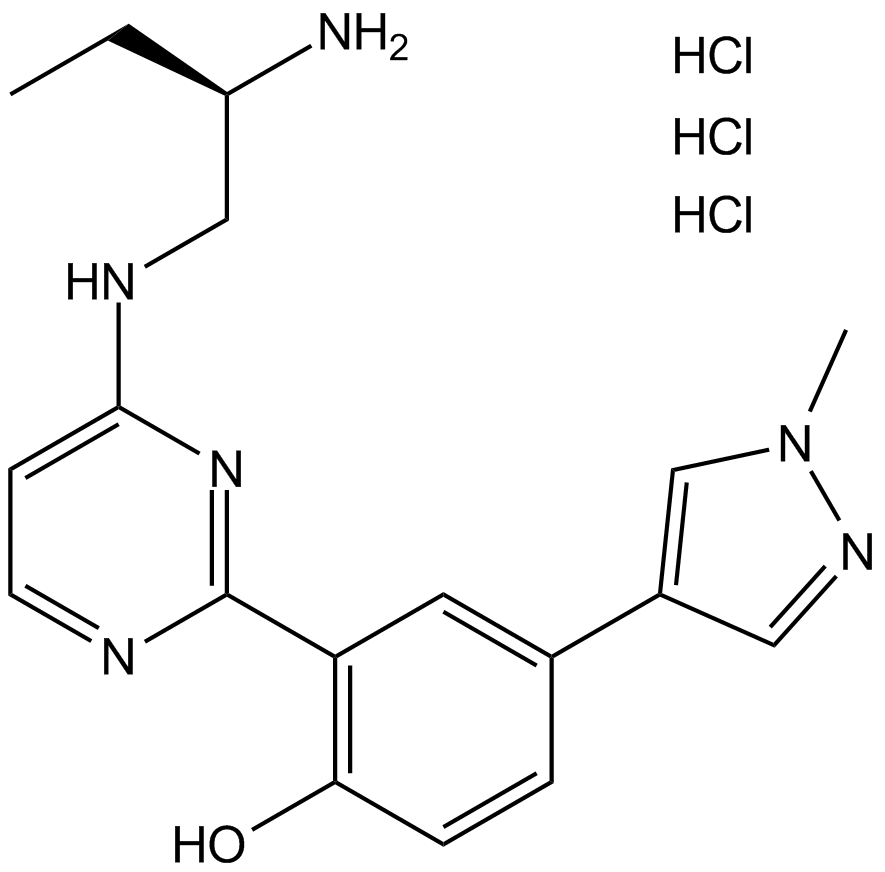
-
GC35750
CRT0066101 dihydrochloride
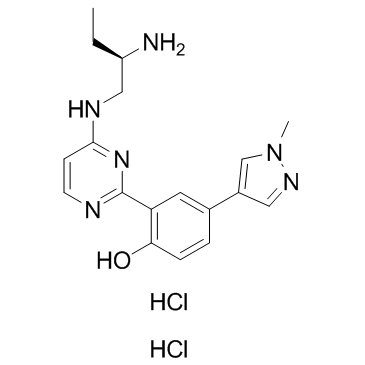
CRT0066101 dihydrochloride is a potent and specific PKD inhibitor with IC50 values of 1, 2.5 and 2 nM for PKD1, 2, and 3 respectively.
-
GC45414
CRT0066854

-
GC14355
CRT5
CRT0066051
PKD1, PKD2, and PKD3 inhibitor
-
GC32911
CTX1
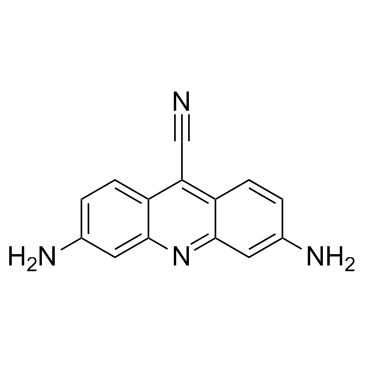
CTX1 is a p53 activator that overcomes HdmX-mediated p53 repression. CTX1 exhibits potent anti-cancer activity in a mouse acute myeloid leukemia (AML) model system.
-
GN10535
Cucurbitacin B
Cuc B, NSC 49451, NSC 144154

-
GC35758
Cucurbitacin IIa
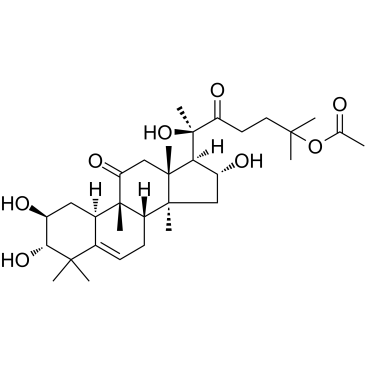
Cucurbitacin IIa is a triterpene isolated from Hemsleya amalils Diels, induces apoptosis of cancer cells, reduces expression of survivin, reduces phospho-Histone H3 and increases cleaved PARP in cancer cells.
-
GN10788
Cucurbitacin IIb
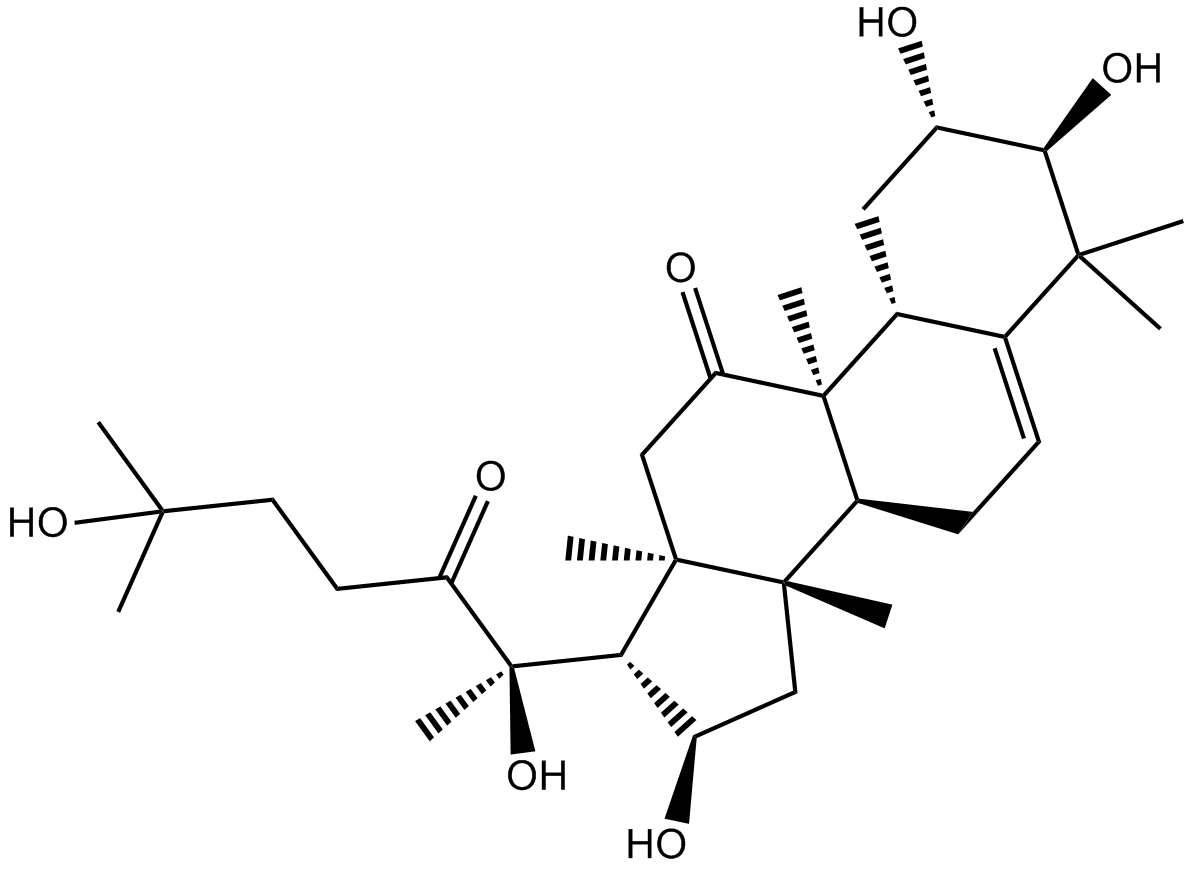
Cuc B, NSC 49451, NSC 144154
-
GC32781
CUDC-427 (GDC-0917)
GDC-0917
CUDC-427 (GDC-0917) is a potent second-generation pan-selective IAP antagonist, used for treatment of various cancers.
-
GC12115
CUDC-907
CUDC-907
CUDC is an orally bioavailable small molecule PI3K and HDAC dual inhibitor that acts on PI3K α And HDAC1 / 2 / 3 / 10 with IC50 of 19 nm and 1.7 nm / 5 nm / 1.8 nm / 2.8 nm.
-
GC11217
CUR 61414
G-856
potent inhibitor of hedgehog-induced cellular activity
-
GC14787
Curcumin
Indian Saffron, Turmeric yellow
A yellow pigment with diverse biological activities
-
GC40226
Curcumin-d6
Curcumin-d6 is intended for use as an internal standard for the quantification of curcumin by GC- or LC-MS.

-
GN10521
Curcumol
(-)-Curcumol



