Alzheimer
Alzheimer
Alzheimer’s disease (AD), a neurodegenerative disorder, is the most common cause of progressive dementia. Two microscopic characteristics of AD are extracellular amyloid plaques and intracellular neurofibrillary tangles. Amyloid β peptide (Aβ), derived from amyloid precursor protein (APP) by sequential protein cleavage, and other metabolites deposit around neurons and form amyloid plaques, which contribute to the disease’s pathogenesis. The neurofibrillary tangles are formed by the aggregation of phosphorylated tau proteins. Under pathogenic conditions, tau accumulates in dendritic spines and interferes with neurotransmission. The Aβoligomer promotes tau enrichment and facilitates disease progress.
Products for Alzheimer
- Cat.No. Product Name Information
-
GC11965
(±)-Huperzine A
Hup A, (-)-Selagine
A neuroprotective AChE inhibitor
-
GC26199
1-heptadecanoyl-2-hydroxy-sn-glycero-3-phosphocholine

-
GC46451
16F16
A PDI inhibitor

-
GC65195
2-O-Acetyl-20-hydroxyecdysone
β-Ecdysone 2-acetate
2-O-Acetyl-20-hydroxyecdysone, an ecdysterones in insects and terrestrial plants, inhibits amyloid-β42 (Aβ42)-induced cytotoxicity.
-
GC18817
3β-hydroxy-5-Cholestenoic Acid
3β-hydroxy-5-CA, Cholesterol-26-carboxylic Acid
3β-hydroxy-5-Cholestenoic acid is an active metabolite of cholesterol formed when cholesterol is metabolized by the cytochrome P450 (CYP) isomer CYP27A1.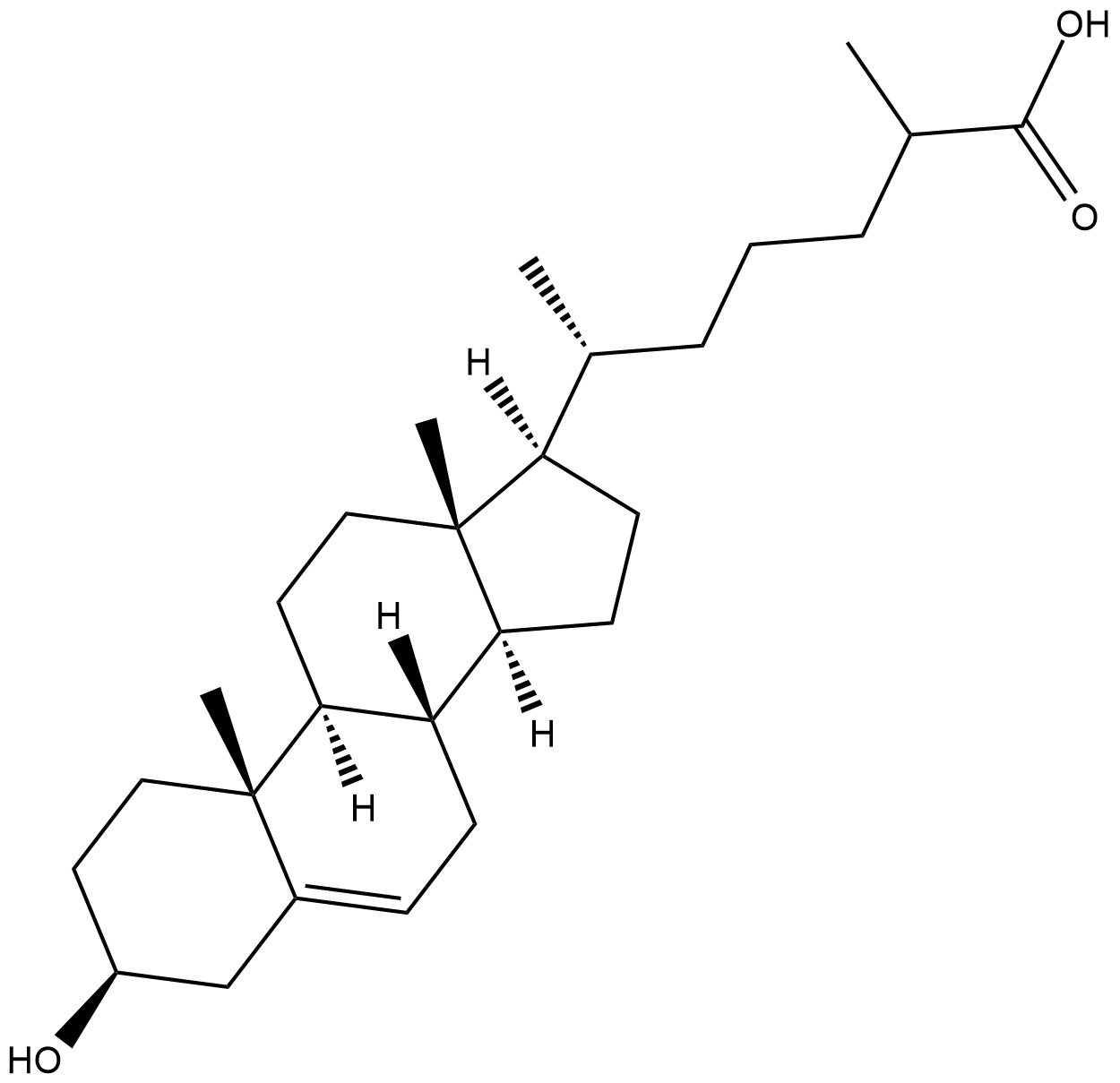
-
GC40053
5α,6α-epoxy Cholestanol
NSC 18176
An oxysterol and a metabolite of cholesterol produced by oxidation
-
GC52172
5-hydroxy Indole-3-acetic Acid-d6
5-HIAA-d6, 5-Hydroxyindoleacetic Acid-d6, 5-hydroxy IAA-d6

-
GC46720
6,9-Dichloro-1,2,3,4-tetrahydroacridine
NSC 1227
A synthetic intermediate in the synthesis of AChE inhibitors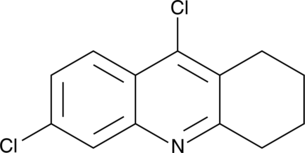
-
GC42584
6-O-desmethyl Donepezil
6-O-desmethyl Donepezil is an active metabolite of the acetylcholinesterase inhibitor donepezil.

-
GC40587
8,12-iso-iPF2α-VI
8,12-iso-Isoprostane-F2α-VI, 12-iso-5,6E,14Z-PGF2α, 12iso5,6E,14ZProstaglandin F2α
8,12-iso-iPF2α-VI is an isoprostane produced by non-enzymatic, free radical-induced peroxidative damage to membrane lipids.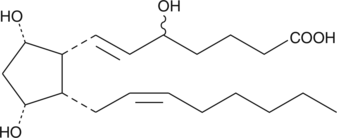
-
GC45378
Alaproclate (hydrochloride)
GEA 654, A03
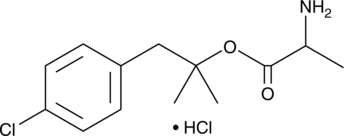
-
GC40024
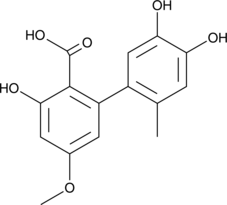
Altenusin
Alutenusin
Altenusin is a polyphenol fungal metabolite originally isolated from the fungus Alternaria that has diverse biological activities.
-
GC42794
Amylin (8-37) (human) (trifluoroacetate salt)
IAPP (8-37) (human), Islet Amyloid Polypeptide (8-37) (human)
Amylin (8-37) is a peptide fragment of amylin.
-
GC42795
Amylin (human) (amidated) (trifluoroacetate salt)
IAPP (human) (amidated), Islet Amyloid Polypeptide (human) (amidated)
Amylin is a peptide hormone secreted from pancreatic β-cells that reduces food intake, decreases glucagon secretion, slows gastric emptying, and increases satiety.
-
GC42796
Amylin (human) (trifluoroacetate salt)
IAPP (human), Islet Amyloid Polypeptide (human)
Amylin is a 37-residue peptide hormone secreted from pancreatic β-cells that reduces food intake, decreases glucagon secretion, slows gastric emptying, and increases satiety.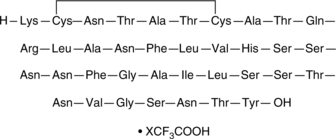
-
GP10099
amyloid A protein fragment [Homo sapiens]
H2N-Ala-Gly-Leu-Pro-Glu-Lys-Tyr-OH
Apolipoproteins related to HDL in plasma![amyloid A protein fragment [Homo sapiens] Chemical Structure amyloid A protein fragment [Homo sapiens] Chemical Structure](/media/struct/GP1/GP10099.png)
-
GP10118
Amyloid Beta-Peptide (1-40) (human)
Amyloid β-Peptide (1-40) (human), (C194H295N53O58S1), a peptide with the sequence H2N-DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA-OH, MW= 4329.8. Amyloid beta (Aβ or Abeta) is a peptide of 36–43 amino acids that is processed from the Amyloid precursor protein.

-
GP10049
Amyloid Beta-Peptide (12-28) (human)
Amyloid Beta-Peptide (12-28) (human) is a peptide fragment of amyloid beta protein (1-42) (Aβ (1-42)).
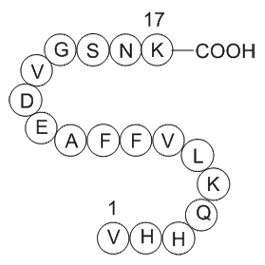
-
GP10082
Amyloid Beta-peptide (25-35) (human)
Amyloid beta(Aβ)-peptide (25-35) (human) is an fragment of Alzheimer's Amyloid beta peptide which is commonly found in the brains of people with Alzheimer's disease (AD) and is a major component of Alzheimer's amyloid plaques.

-
GP10046
Amyloid Precursor C-Terminal Peptide
H2N-Gly-Tyr-Glu-Asn-Pro-Thr-Tyr-Lys-Phe-Phe-Glu-Gln-Met-Gln-Asn-OH
For beta amyloid generation

-
GP10057
Amyloid β-Peptide (10-20) (human)
Tyr-Glu-Val-His-His-Gln-Lys-Leu-Val-Phe-Phe

-
GP10094
Amyloid β-peptide (10-35), amide
H2N-Tyr-Glu-Val-His-His-Gln-Lys-Leu-Val-Phe-Phe-Ala-Glu-Asp-Val-Gly-Ser-Asn-Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met-NH2

-
GP10097
Amyloid β-Protein (1-15)
H2N-Asp-Ala-Glu-Phe-Arg-His-Asp-Ser-Gly-Tyr-Glu-Val-His-His-Gln-OH
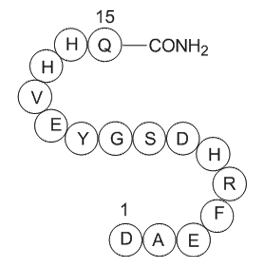
-
GC46851
Amyloid-β (1-42) Peptide (trifluoroacetate salt)
Aβ (1-42), Aβ42
A 42-amino acid protein fragment of amyloid-β
-
GC42801
Amyloid-β (1-8) Peptide
Aβ (1-8)
Amyloid-β (1-8) is a wild-type control for the mutation-containing amyloid-β (1-8, A2V) peptide .
-
GC42802
Amyloid-β (1-8, A2V) Peptide
Aβ (1-8, A2V), Aβ (1-8) mutant, Amyloid-β (1-8) dominant negative
Amyloid-β (1-8, A2V) is a truncated form of amyloid-β (Aβ) that contains a valine to alanine substitution at position 2 of the Aβ numbering convention (Aβ A2V), which corresponds to position 673 of the amyloid precursor protein (APP) numbering convention (APP A673V).
-
GC42803
Amyloid-β (25-35) Peptide (human) (trifluoroacetate salt)
Aβ (25-35)
Amyloid-β (25-35) (Aβ (25-35)) is an 11-residue fragment of the Aβ protein that retains the physical and biological characteristics of the full length peptide.
-
GP10083
Beta-Amyloid (1-11)
H2N-Asp-Ala-Glu-Phe-Arg-His-Asp-Ser-Gly-Tyr-Glu-OH
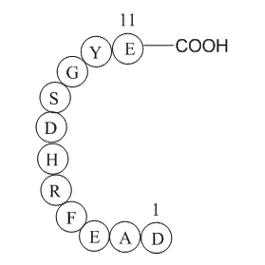
-
GP10101
Beta-Sheet Breaker Peptide iAβ5
H2N-Leu-Pro-Phe-Phe-Asp-OH

-
GC42937
Biotin-Amyloid-β (1-42) Peptide (trifluoroacetate salt)
Biotin-Aβ (1-42), Biotin-Aβ42
Biotin-amyloid-β (1-42) peptide is an affinity probe that allows amyloid-β (1-42) (Aβ42) to be detected or immobilized through interaction with the biotin ligand.
-
GC46992
C18 Ganglioside GM1-d3 (d18:1/18:0-d3) (ammonium salt)
N-omega-CD3-Octadecanoyl monosialoganglioside GM1-d3, N-CD3-Stearoyl-GM, C18 GM1-d3, GM1 (18:1/18:0-d3), GM1 (d18:1/C18:0)-d3, Monosialoganglioside GM1-d3, N-Stearoyl Monosialoganglioside GM1-d3
A neuropeptide with diverse biological activities
-
GC46997
C22 Galactosylceramide (d18:1/22:0)
N-Docosanoyl-β-D-Galactosylceramide, Galactosylceramide (d18:1/22:0), GalCer(d18:1/22:0)
A sphingolipid
-
GC46998
C22 Galactosylceramide-d4 (d18:1/22:0-d4)
Galactosylceramide-d4 (d18:1/22:0-d4), N-Docosanoyl-β-D-Galactosylceramide-d4, GalCer(d18:1/22:0-d4)
A neuropeptide with diverse biological activities
-
GC91899
Carmustine-d8
BCNU-d8; bis-Chloroethylnitrosourea-d8
Carmustine-d8 is intended for use as an internal standard for the quantification of carmustine by GC- or LC-MS.
-
GC43274
Citromycetin
NSC 53584
Citromycetin is a fungal metabolite originally isolated from P.
-
GP10010
COG 133
Leu-Arg-Val-Arg-Leu-Ala-Ser-His-Leu-Arg-Lys-Leu-Arg-Lys-Arg-Leu-Leu

-
GC43304
Compound 43 TAO Kinase Inhibitor
Compound 43 TAO kinase inhibitor is an ATP-competitive inhibitor of the thousand-and-one amino acid kinases TAOK1 and TAOK2 (IC50s = 11 and 15 nM, respectively).

-
GC43322
CRANAD 28
CRANAD 28 is a curcumin derivative and non-conjugated fluorescent affinity probe for the detection of amyloid-β (Aβ) in vitro and in vivo that has excitation/emission spectra of 498/578 nm, respectively.

-
GC49557
Cu-GTSM
CuII(gtsm), copper-GTSM
A copper-containing compound with diverse biological activities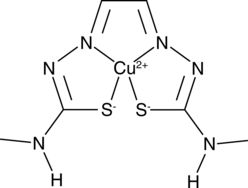
-
GC12942
DAPT (GSI-IX)
gamma-Secretase Inhibitor IX, DAPT, GSI-IX
DAPT (GSI-IX) is an orally active γ-secretase inhibitor with IC50s of 115 nM and 200 nM for total amyloid-β (Aβ) and Aβ42 produced in human primary neuronal cultures, respectively.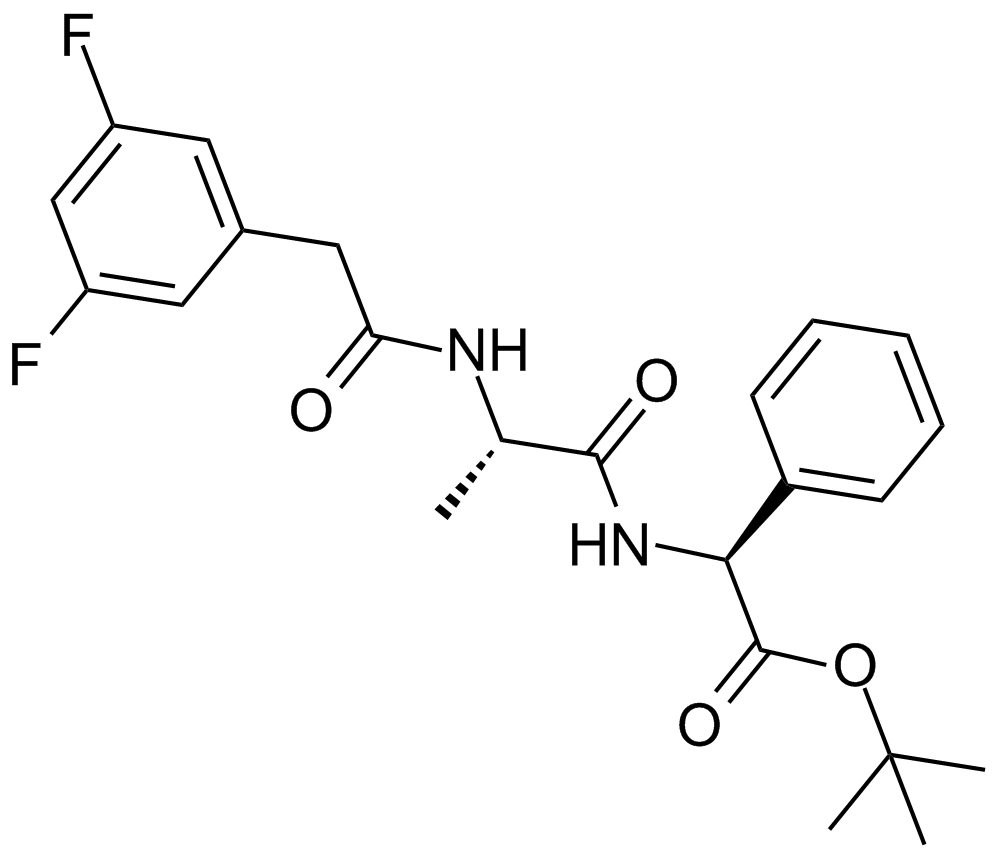
-
GC49913
Davunetide (acetate)
AL-108, NAP, NAPVSIPQ
A neuroprotective ADNP-derived peptide
-
GC41151
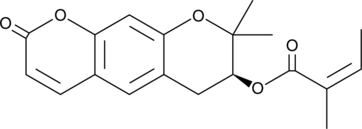
Decursinol angelate
Decursinol angelate is a pyranocoumarin that has been found in the Korean medicinal herb A.
-
GC45765
Deferiprone-d3
An internal standard for the quantification of deferiprone

-
GC47186
Deoxy Donepezil (hydrochloride)
A potential impurity found in bulk preparations of donepezil

-
GC47196
Desloratadine-d4
Descarboethoxyloratadine-d4
A neuropeptide with diverse biological activities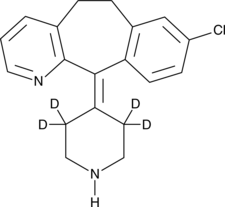
-
GC46134
Dityrosine (hydrochloride)
Bityrosine, o,o-Ditryosine
A neuropeptide with diverse biological activities
-
GC43494
DL-threo-PDMP (hydrochloride)
DL-threo PDMP is a mixture of ceramide analogs that contains two of the four possible stereoisomers of PDMP : D-threo (1R,2R) and L-threo (1S,2S) PDMP.

-
GC47263
Donecopride (fumarate hydrate)
MR31147
A 5-HT4 receptor partial agonist and AChE inhibitor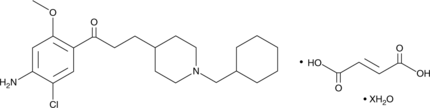
-
GC40018
Donepezil N-oxide
Donepezil N-oxide is an active metabolite of the acetylcholinesterase inhibitor donepezil.

-
GC47264
Donepezil-d4 (hydrochloride)
An internal standard for the quantification of donepezil

-
GC43700
FSB
FSB is a fluorescent probe that has been used for the detection of amyloid plaques in human postmortem brain from patients with Alzheimer's disease and of curli amyloid fibers in E.

-
GC43708
Fulvic Acid
Fulvic acid is a phenolic acid and fungal metabolite originally isolated from Penicillium.

-
GC52047
Fustin
Dihydrofisetin, NSC 59264
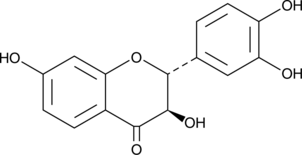
-
GC52078
Galantamine (hydrobromide)
Galanthamine, NSC 100058
An alkaloid with AChE inhibitory and nAChR potentiating activities
-
GC47389
Galantamine-d3 (hydrobromide)
An internal standard for the quantification of galantamine

-
GC20007
Ginsenoside CK
Ginsenoside Compound K, Ginsenoside IH901
A metabolite of ginsenoside Rb1 with diverse biological activity
-
GC49273
Glycerophosphorylethanolamine (sodium salt)
Glycerol 3-phosphorylethanolamine, Glycerylphosphorylethanolamine
An active metabolite of phosphatidylethanolamine
-
GC40114
Histone H3K4Me3 (1-10) (human, mouse, rat, porcine, bovine) (trifluoroacetate salt)
Histone H3K4Me3 (1-10) is an N-terminal peptide fragment of histone H3 that corresponds to amino acid residues 2-11 of the human histone H3 sequence.

-
GC43858
Histone H3K4Me3 (1-21) (human, mouse, rat, porcine, bovine) (trifluoroacetate salt)
ART-K(Me3)-QTARKSTGGKAPRKQLA, H3K4(Me3), Histone H3 (1-21) (Lys4me3), Lys(Me3)4-Histone H3 (1-21)
Histone H3K4Me3 (1-21) is an N-terminal peptide fragment of histone H3 that corresponds to amino acid residues 2-22 of the human histone H3 sequence.

-
GC45480
Histone H3K4Me3 (1-25) amide (human mouse, rat, porcine, bovine) (trifluoroacetate salt)
ART-K(Me3)-QTARKSTGGKAPRKQLATKAA-NH2, Histone H3 (1-25) (Lys4me3), H3K4me3, Lys(Me3)4-Histone H3 (1-25) amide

-
GC47437
Humanin (human) (trifluoroacetate salt)
HN
A neuropeptide with diverse biological activities
-
GC49493
IRL 1620 (trifluoroacetate salt)
PMZ-1620, SPI 1620, Suc-Glu9, Ala11,15-ET-1(8-21)
A peptide ETB receptor agonist
-
GC52027
Isolariciresinol
(+)-Cyclolariciresinol, (+)-Isolariciresinol, (7′S,8′R,8R)-Isolariciresinol
A lignan with diverse biological activities
-
GC49306
Isopimaric Acid
7,15-Isopimaradien-18-oic Acid, (+)-Isopimaric Acid
A diterpenoid resin acid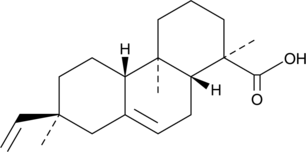
-
GC18568
JNJ-40418677
A modulator of γ-secretase

-
GC49436
L-3-n-Butylphthalide
(-)-3-Butylphthalide, levo-3-n-Butylphthalide, (S)-(-)-3-Butylphthalide, L-NBP
A phthalide with antiplatelet and neuroprotective activities
-
GC47563
L-Glutamic Acid (ammonium salt)
Glutamic Acid
An excitatory neurotransmitter
-
GC49417
L-Serine-13C3
(S)-Serine-13C3
An internal standard for the quantification of L-serine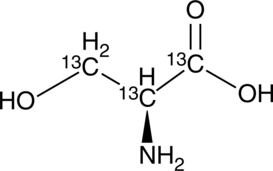
-
GC49461
L-Serine-d7
(S)-Serine-d7
An internal standard for the quantification of L-serine
-
GC45917
Leucettine L41
An inhibitor of DYRK1A, DYRK2, CLK1, and CLK3

-
GC19221
Leucomethylene blue Mesylate
Hydromethylthionine, Leucomethylene Blue, Leukomethylthioninium dihydromesylate, LMTM
Leucomethylene blue (Mesylate) is a common reduced form of Methylene Blue, Methylene Blue is a member of the thiazine class of dyes.

-
GC44130
Mca-EVKMDAEF-K(Dnp)-NH2 (ammonium salt)
β-Secretase Fluorogenic Substrate
Mca-EVKMDAEF-K(Dnp)-NH2 is a fluorogenic substrate for β-secretase.
-
GC44131
Mca-HQKLVFFA-K(Dnp)-NH2 (trifluoroacetate salt)
α-Secretase Fluorogenic Substrate
Mca-HQKLVFFA-K(Dnp)-NH2 is a fluorogenic substrate for α-secretase.
-
GC40078
Memantine-d6 (hydrochloride)
Memantine-d6 is intended for use as an internal standard for the quantification of memantine by GC- or LC-MS.

-
GC44225
ML-345
Insulin-degrading enzyme (IDE) is a thiol-sensitive zinc-metallopeptidase that acts as the major insulin-degrading protease in vivo, mediating the termination of insulin signaling.

-
GP10006
Myelin Basic Protein (68-82), guinea pig

-
GP10130
Myelin Basic Protein (87-99)
Val-His-Phe-Phe-Lys-Asn-Ile-Val-Thr-Pro-Arg-Thr-Pro
An encephalitogenic peptide

-
GC44416
N-methyl Mesoporphyrin IX
N-methyl Mesoporphyrin IX (NMM) is a transition state analog of porphyrin and an effective inhibitor of ferrochelatase with a Kd value of 7nM. N-methyl Mesoporphyrin IX is a specific stain for G-quadruplexes, with excitation/emission maxima at 399/610 nm.

-
GC47746
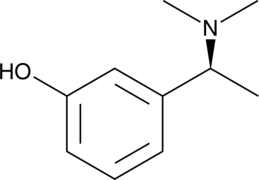
NAP 226-90
A metabolite of rivastigmine
-
GC44399
NIAD-4
NIAD-4 is a fluorescent probe that crosses the blood-brain barrier to bind with high affinity to amyloid-β (Aβ) plaques (Ki = 10 nM).

-
GC52214
Nicotinamide riboside-d4 (triflate)
An internal standard for the quantification of nicotinamide riboside

-
GC49090
Nilvadipine-d4
(±)-Nilvadipine-d4
An internal standard for the quantification of nilvadipine
-
GC72054
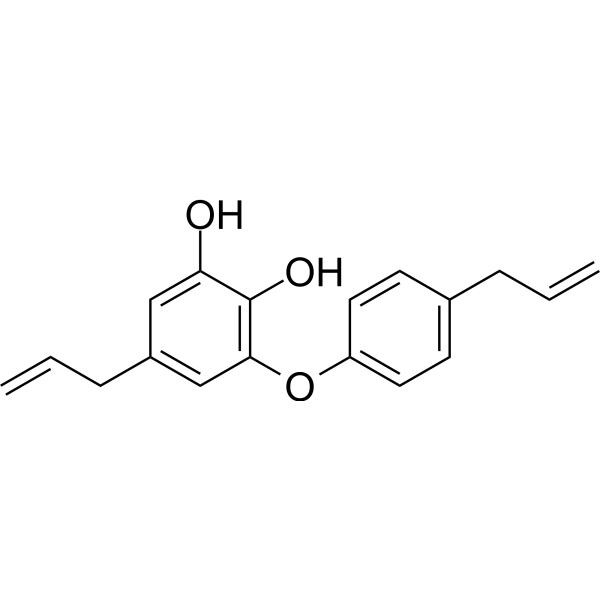
Obovatol
Obovatol is a biphenyl ether lignan isolated from the leaves of Magnolia obovata Thunb.
-
GC49815
Oleuropein aglycone
3,4-DHPEA-EA
A polyphenol with diverse biological activities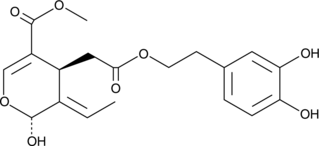
-
GC91070
p-methyl-N-salicyloyl Tryptamine
A neuroprotective agent

-
GC49024
Palmitic Acid MaxSpec® Standard
C16:0, Cetylic Acid, NSC 5030
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
GC49023
Palmitic Acid-d9 MaxSpec® Standard
C16:0-d9, Cetylic Acid-d9
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
GC39419
PBD-150
PBD-150 is a human glutaminyl cyclase (hQC) Y115E-Y117E variant inhibitor, with a Ki value of 490 nM.

-
GC47955
Phloroglucinol
Benzene-s-triol, 1,3,5-Hydroxybenzene, NSC 1572, 1,3,5-Trihydroxybenzene, sym-Trihydroxybenzene
A phenol with diverse biological activities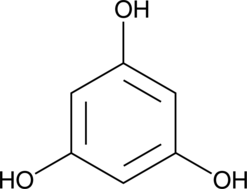
-
GC49192
Piracetam-d6
UCB-6215-d6
An internal standard for the quantification of piracetam
-
GC48362
PMX-205 (trifluoroacetate salt)
A potent antagonist of C5aR

-
GC44674
Pramlintide (acetate hydrate)
Pramlintide is a non-amyloidogenic analog of the antidiabetic peptide hormone amylin that contains proline residues substituted at positions 25, 28, and 29.

-
GC46208
Propentofylline
HOE 285, HWA 285
A xanthine derivative and neuroprotective agent
-
GC49108
Racecadotril-d5
Acetorphan-d5
An internal standard for the quantification of racecadotril
-
GC48028
Rasagiline-13C3 (mesylate)
An internal standard for the quantification of rasagiline

-
GC18483
RU-505
RU-505 is an inhibitor of the interaction between amyloid-β (Aβ) and fibrinogen, with a higher efficacy for inhibiting monomeric forms of Aβ bound to fibrinogen over oligomeric forms.

-
GC19799
Selegiline hydrochloride
R(-)-Deprenyl hydrochloride;R(-)-Deprenyl hydrochloride
Selegiline hydrochloride is a selective, reversible inhibitor of MAO-B (Ki = 0.091 μM) over MAO-A (Ki = 9.06 μM). Monoamine oxidase (MAO) inhibitors have utility in ameliorating a variety of neurological conditions, including depression.

-
GC12096
Semagacestat (LY450139)
LY450139
A pan γ-secretase inhibitor
-
GC40068
Setosusin
(+)-Setosusin
Setosusin is a meroterpenoid fungal metabolite originally isolated from C.
-
GC48083
sn-Glycerol-3-phosphate (cyclohexyl ammonium salt hydrate)
G3P
A neuropeptide with diverse biological activities



