STING
STING (Stimulator of Interferon Genes) is a transmembrane protein localized to the endoplasmic reticulum that undergoes a conformational change in response to direct binding of cyclic dinucleotides (CDNs), resulting in a downstream signaling cascade involving TBK1 activation, IRF-3 phosphorylation, and production of IFN-β and other cytokines.
STING is a pattern recognition receptor of cyclic dinucleotides as well as an innate immune adaptor protein that enables signaling from cytoplasmic receptors to the transcription factor interferon regulatory factor 3. Initiation of these pathways leads to the expression of type I interferons and proteins associated with antiviral and antitumor immunity. Small molecules capable of triggering STING-dependent cellular processes are effective at blocking virus replication, enhancing vaccine efficacy, and facilitating immune response to cancer cells.
Targets for STING
Products for STING
- Cat.No. Product Name Information
-
GC49912
13C20,15N10-Cyclic di-GMP (sodium salt)
13C20,15N10-c-di-GMP, 13C20,15N10-Cyclic diguanylate, 13C20,15N10-3’,5’-Cyclic diguanylic Acid
An internal standard for the quantification of cyclic di-GMP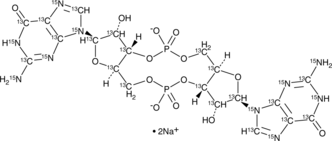
-
GC42080
2'2'-cGAMP (sodium salt)
Adenosine-Guanosine 2’,2’-cyclic monophosphate, cGAMP(2’-5’), 2’,2’-Cyclic GMP-AMP
2'2'-cGAMP is a synthetic dinucleotide (CDN) that contains non-canonical 2'5'-phosphodiester bonds.
-
GC42090
2'3'-cGAMP (sodium salt)
Guanosine-Adenosine 2',3'-cyclic monophosphate, 2'-3'-Cyclic GMP-AMP
2'3'-cGAMP is a second messenger produced from ATP and GTP by cGMP-AMP synthase (cGAS) in the cytoplasm of mammalian cells in response to the presence of DNA.

-
GC25014
3',3'-cGAMP
3',3'-cyclic GMP-AMP, Cyclic GMP-AMP, cGAMP
3',3'-cGAMP (3',3'-cyclic GMP-AMP, Cyclic GMP-AMP, cGAMP) activates the endoplasmic reticulum (ER)-resident receptor stimulator of interferon genes (STING), thereby inducing an antiviral state and the secretion of type I IFNs.
-
GC42245
3'3'-cGAMP (sodium salt)
AdenosineGuanosine 3',3'-cyclic monophosphate, 3',3'-Cyclic GMP-AMP, c-GpAp
3'3'-cGAMP is a second messenger produced in bacteria by specific dinucleotide cyclases.
-
GC48381
5'-pApA (sodium salt)
c-di-AMP Control, Cyclic di-AMP Negative Control
A linearized form of cyclic di-AMP
-
GC48375
5'-pGpG (sodium salt)
c-di-GMP Control, Cyclic di-GMP Negative Control
A linearized form of cyclic di-GMP
-
GC49238
93-O17S
A cationic lipidoid

-
GC31649
ADU-S100 (ML RR-S2 CDA)
MIW815; ML RR-S2 CDA
ADU-S100 (ML RR-S2 CDA) (MIW815), an activator of stimulator of interferon genes (STING), leads to potent and systemic tumor regression and immunity.
-
GC39161
ADU-S100 disodium salt
MIW815 disodium salt; ML RR-S2 CDA disodium salt
ADU-S100 disodium salt (MIW815 disodium salt) is an activator of stimulatory interferon gene (STING) protein with a Kd of 4.61±0.42μM[1].
-
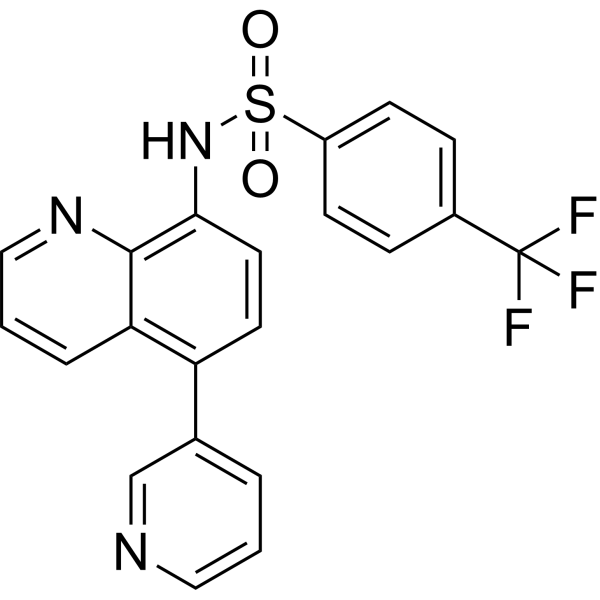
GC74005
AK59
AK59 is a STING degrader that works by leveraging HERC4, a hect domain E3 ligase.
-
GC71433
Anti-inflammatory agent 70
Anti-inflammatory agent 70 (N-Me-SP23) is a STING protein degrader and inhibits the STING signaling pathway.

-
GC42880
Avenanthramide-C methyl ester
Avenanthramide-C methyl ester is an inhibitor of NF-κB activation that acts by blocking the phosphorylation of IKK and IκB (IC50 ~ 40 μM).

-
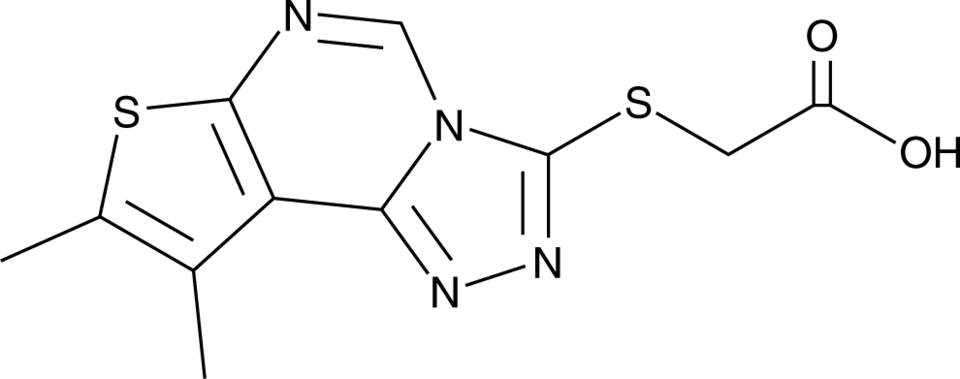
GC91659
BDW-OH
BDW-OH is an active metabolite of the STING agonist and prodrug BDW568 .
-
GC91379
BDW568
BDW568 is an agonist of stimulator of interferon genes (STING) and a prodrug form of BDW-OH.

-
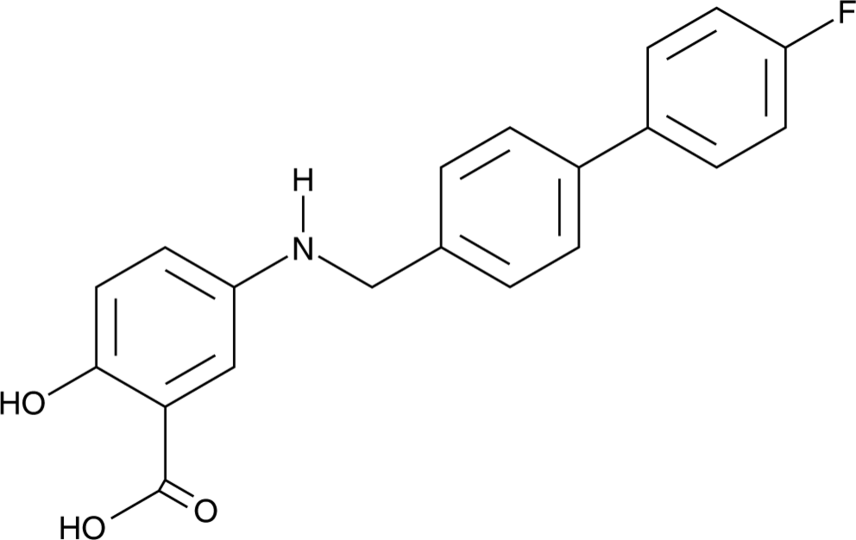
GC91947
BH400
BH400 is an agonist of peroxisome proliferator-activated receptor α (PPARα) and an inhibitor of stimulator of interferon genes (STING; IC50 = 8.1 ?M).
-
GC19069
BI605906
BI605906 is a novel IKKβ inhibitor with an IC50 value of 380 nM when assayed at 0.1 mM ATP.

-
GC42953
BMS 345541 (trifluoroacetate salt)
BMS 345541 is a cell permeable inhibitor of the IκB kinases IKKα and IKKβ (IC50s = 4 and 0.3 μM).
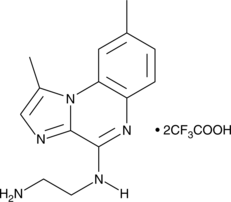
-
GC90705
BSP-16
A STING agonist
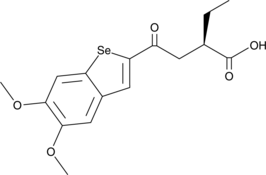
-
GC46983
C-170
C-170 (C-170) is a potent and covalent STING inhibitor.
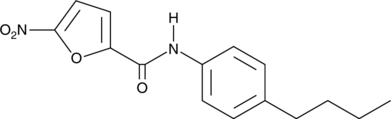
-
GC46984
C-171
C-171 is an inhibitor of stimulator of interferon genes (STING).

-
GC33823
C-176 (STING inhibitor 1)
C-176 (STING inhibitor 1) strongly reduces STING-mediated, but not RIG-I- or TBK1-mediated, IFNβ reporter activity.
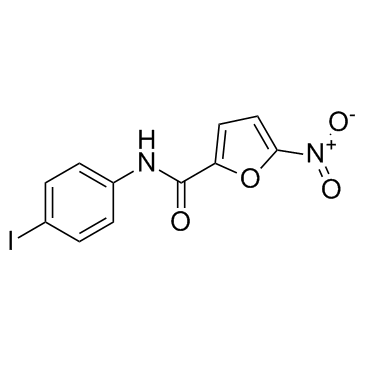
-
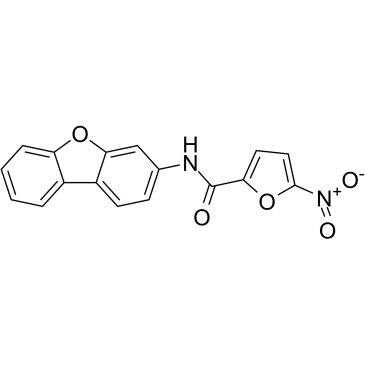
GC38494
C-178
A STING inhibitor
-
GC40570
C-178
C-178 is a covalent inhibitor of STING.

-
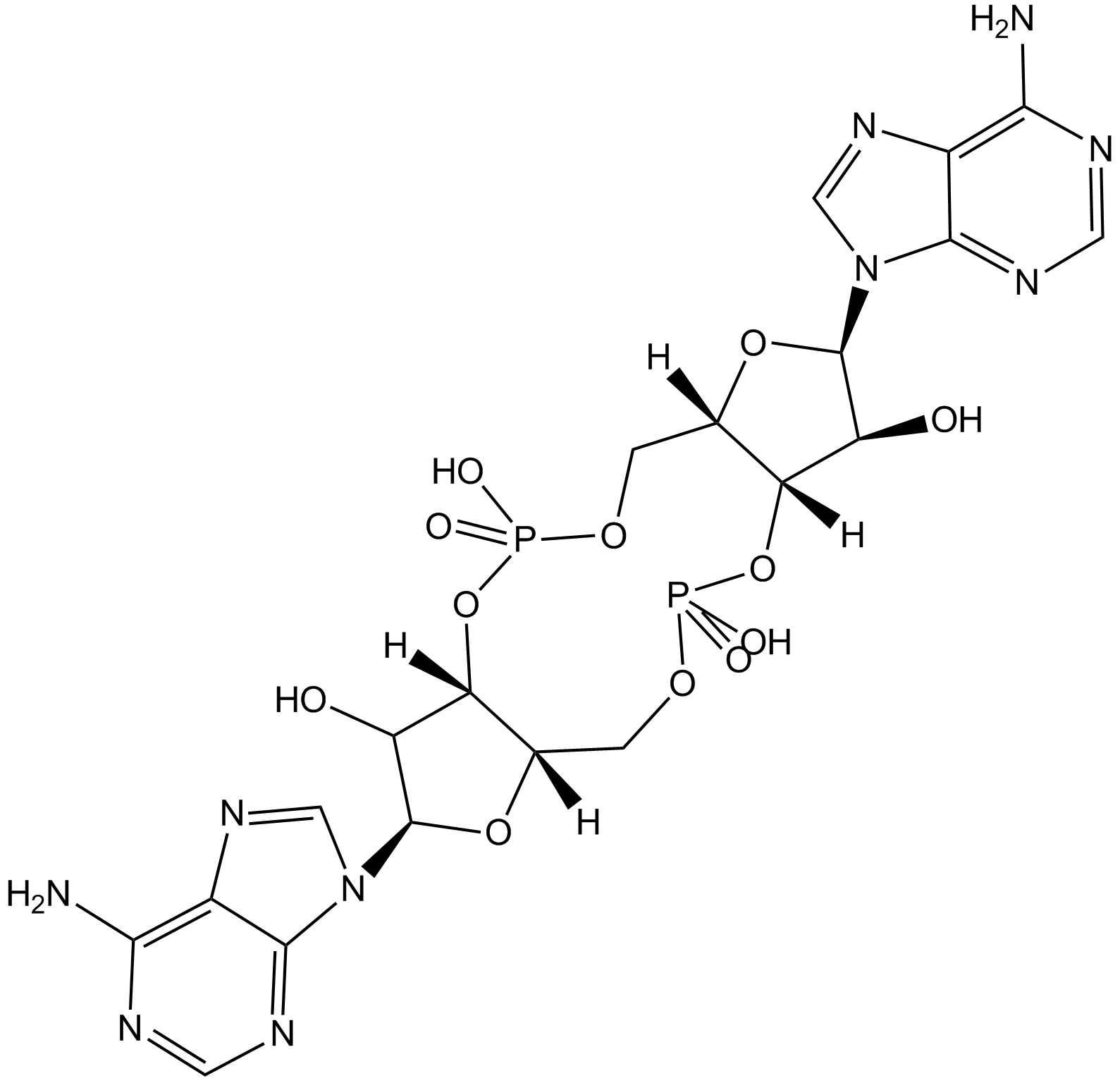
GC13643
c-di-AMP
C-di-AMP is a STING agonist, which binds to the transmembrane protein STING, thereby activating the TBK3-IRF3 signaling pathway, subsequently triggering the production of type I IFN and TNF.
-
GC62893
c-di-AMP diammonium
Cyclic diadenylate diammonium; Cyclic-di-AMP diammonium
c-di-AMP diammonium is a STING agonist, which binds to the transmembrane protein STING thereby activating the TBK3-IRF3 signaling pathway, subsequently triggering the production of type I IFN and TNF.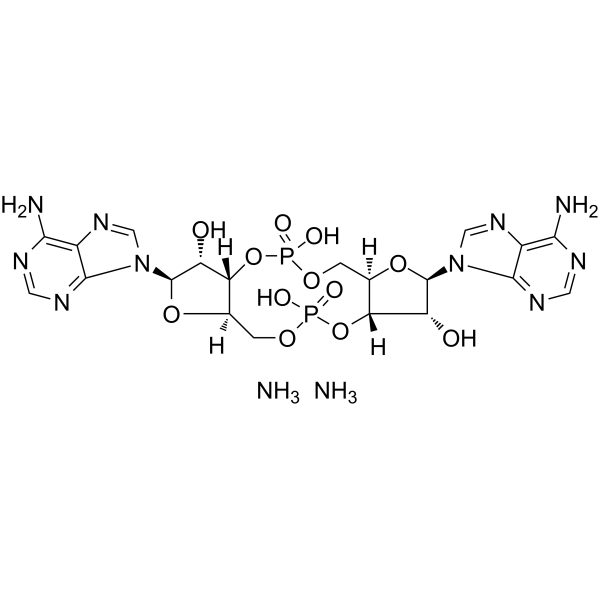
-
GC64445
c-di-AMP disodium
c-di-AMP, Cyclic di-Adenosine monophosphate, 3',5'-Cyclic diadenylic acid, Cyclic diadenylate
c-di-AMP (Cyclic diadenylate) sodium is a STING agonist, which binds to the transmembrane protein STING thereby activating the TBK3-IRF3 signaling pathway, subsequently triggering the production of type I IFN and TNF.
-
GC62198
c-di-AMP sodium
c-di-AMP, Cyclic di-Adenosine monophosphate, 3',5'-Cyclic diadenylic acid, Cyclic diadenylate
C-di-AMP is a STING agonist, which binds to the transmembrane protein STING, thereby activating the TBK3-IRF3 signaling pathway, subsequently triggering the production of type I IFN and TNF.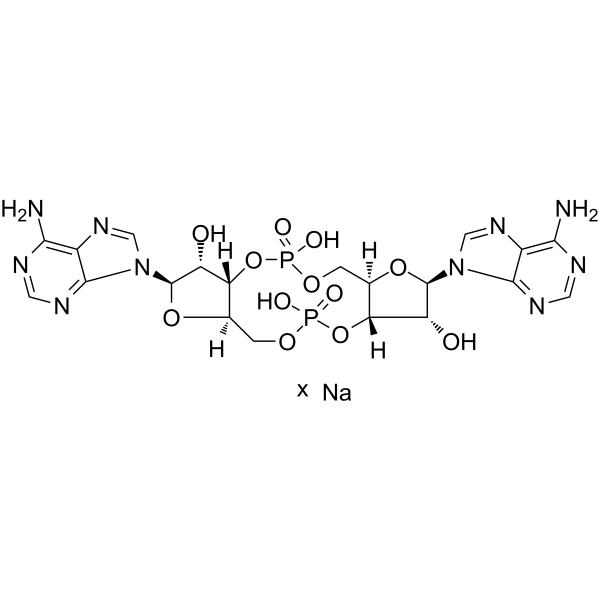
-
GC50687
c-Di-AMP sodium salt

-
GC50292
c-Di-GMP sodium salt
Endogenous STING and DDX41 agonist; activates STING-dependent signaling

-
GC43176
CAY10575
IKK2 Inhibitor 3, Polo-like Kinase Inhibitor 1
CAY10575 (Compound 8) is an IKK2 inhibitor with an IC50 of 0.075 μM.
-
GC18782
CAY10657
The NF-κB/Rel family of transcription factors induce and coordinate the expression of pro-inflammatory genes including cytokines, chemokines, interferons, MHC proteins, growth factors, and cell adhesion molecules.

-
GC47054
CAY10748
A STING agonist
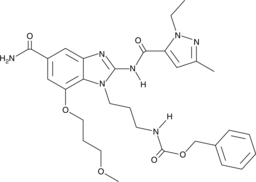
-
GC14727
CCCP
Carbonyl cyanide m-chlorophenyl hydrazone, NSC 88124
Carbonylcyanide-3-chlorophenylhydrazone (CCCP) is a protonophore, which causes uncoupling of proton gradient in the inner mitochondrial membrane, thus inhibiting the rate of ATP synthesis.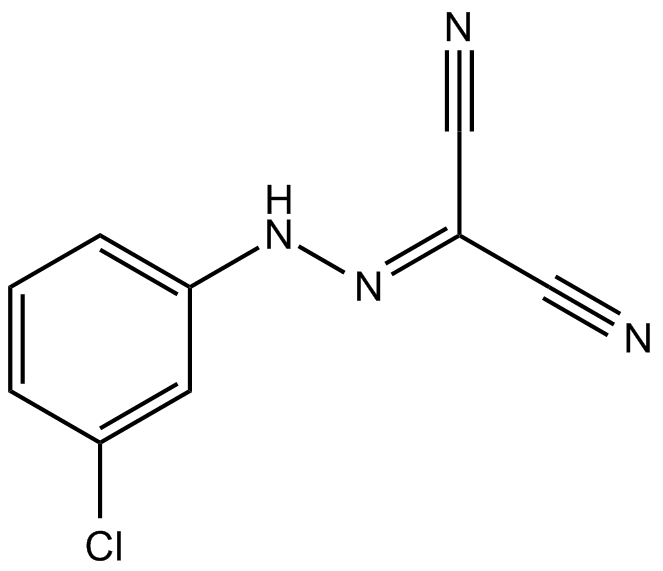
-
GC31696
cGAMP (Cyclic AMP-GMP)
Cyclic GMP-AMP; 3',3'-cGAMP
cGAMP (Cyclic AMP-GMP) (Cyclic GMP-AMPP) functions as an endogenous second messenger in metazoans and triggers interferon production in response to cytosolic DNA.
-
GC63758
cGAMP diammonium
Cyclic GMP-AMP diammonium; 3',3'-cGAMP diammonium
cGAMP (Cyclic GMP-AMPP) diammonium functions as an endogenous second messenger in metazoans and triggers interferon production in response to cytosolic DNA.
-
GC34329
ChX710
ChX710 could prime the type I interferon response to cytosolic DNA, which induces the ISRE promoter sequence, specific cellular Interferon-Stimulated Genes (ISGs), and the phosphorylation of Interferon Regulatory Factor (IRF) 3.

-
GC31875
CL656 (c-[2'FdAMP(S)-2'FdIMP(S)])
c-[2’FdAMP(S)-2’FdIMP(S)]
CL656 (c-[2'FdAMP(S)-2'FdIMP(S)]) is an activator of stimulator of interferon genes (STING).![CL656 (c-[2'FdAMP(S)-2'FdIMP(S)]) Chemical Structure CL656 (c-[2'FdAMP(S)-2'FdIMP(S)]) Chemical Structure](/media/struct/GC3/GC31875.png)
-
GC45413
Cridanimod (sodium salt)
10-Carboxymethyl-9-Acridanone

-
GC47128
CU-32
A cGAS inhibitor

-
GC47129
CU-76
A cGAS inhibitor

-
GC43339
Cyclic di-AMP (sodium salt)
c-di-AMP, Cyclic di-Adenosine monophosphate, 3',5'-Cyclic diadenylic acid, Cyclic diadenylate
Cyclic di-AMP (c-di-AMP) is a second messenger produced by bacteria but not by mammals.
-
GC15048
Cyclic di-GMP
second messenger

-
GC19526
Cyclic di-GMP disodium
c-di-GMP, Cyclic diguanylate, 3',5'-Cyclic diguanylic acid
Cyclic di-GMP (sodium salt) is a STING agonist and a bacterial second messenger that coordinates different aspects of bacterial growth and behavior, including motility, virulence, biofilm formation, and cell cycle progression. Cyclic di-GMP (sodium salt) has anti-cancer cell proliferation activity and also induces elevated CD4 receptor expression and cell cycle arrest. Cyclic di-GMP (sodium salt) can be used in cancer research.
-
GC43340
Cyclic di-IMP (sodium salt)
c-di-IMP, Cyclic di-inosine monophosphate
Cyclic di-IMP (sodium salt) (c-di-IMP) is a synthetic second messenger structurally related to the bacterial second messengers cyclic di-GMP and cyclic di-AMP.
-
GC48397
Cyclic di-UMP (sodium salt)
c-di-UMP, c-UpUp, Cyclic di-Uridine monophosphate, 3’3’-Cyclic UMP-UMP
A pyrimidine-containing CDN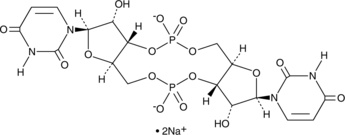
-
GC62917
Cyclic-di-GMP diammonium
c-di-GMP diammonium; cyclic diguanylate diammonium; 5GP-5GP diammonium
Cyclic-di-GMP diammonium is a STING agonist and a bacterial second messenger that coordinates different aspects of bacterial growth and behavior, including motility, virulence, biofilm formation, and cell cycle progression. Cyclic-di-GMP diammonium has anti-cancer cell proliferation activity and also induces elevated CD4 receptor expression and cell cycle arrest. Cyclic-di-GMP diammonium can be used in cancer research.
-
GC62372
Cyclic-di-GMP sodium
c-di-GMP sodium; cyclic diguanylate sodium; 5GP-5GP sodium
Cyclic-di-GMP sodium is a STING agonist and a bacterial second messenger that coordinates different aspects of bacterial growth and behavior, including motility, virulence, biofilm formation, and cell cycle progression. Cyclic-di-GMP sodium has anti-cancer cell proliferation activity and also induces elevated CD4 receptor expression and cell cycle arrest. Cyclic-di-GMP sodium can be used in cancer research.
-
GC35853
diABZI STING agonist-1
diABZI STING agonist-1 is a selective stimulator of interferon genes (STING) receptor agonist, with EC50s of 130, 186 nM for human and mouse, respectively.

-
GC35854
diABZI STING agonist-1 Tautomerism
diABZI STING agonist-1 Tautomerism (compound 3) is a selective stimulator of interferon genes (STING) receptor agonist, with EC50s of 130, 186 nM for human and mouse, respectively.

-
GC35855
diABZI STING agonist-1 trihydrochloride
Diamidobenzimidazole STING Agonist-1, STING Agonist (Compound 3), STING Agonist diABZI
diABZI STING agonist-1 trihydrochloride, as a STING agonist, is internalized into the cytoplasm through unknown receptor and induce the activation and dimerization of STING followed by TBK1/IRF3 phosporylation leading to type I IFN response.
-
GC63473
diABZI-C2-NH2
diABZI-C2-NH2, an active analogue containing a primary amine functionality, is a STING agonist.

-
GC16280
DMXAA (Vadimezan)
AS-1404, 5,6-MeXAA, NSC-640488, Vadimezan
DMXAA (Vadimezan) is a selective DT-diaphorase inhibitor with a Ki value of 20μM and an IC50 value of 62.5μM.

-
GC62608
E7766 diammonium salt
E7766 diammonium salt is a macrocycle-bridged STING agonist with a Kd of 40 nM. E7766 diammonium salt shows potent pan-genotypic and antitumor activities.

-
GC62609
E7766 disodium
E7766 disodium is a macrocycle-bridged STING agonist with a Kd of 40 nM. E7766 disodium shows potent pan-genotypic and antitumor activities.

-
GC73576
Enpp-1-IN-19
Enpp-1-IN-19 (compound 29f) is an orally active ENPP1 inhibitor that inhibits cGAMP drolysis by ENPP1 (IC50=68 nM).
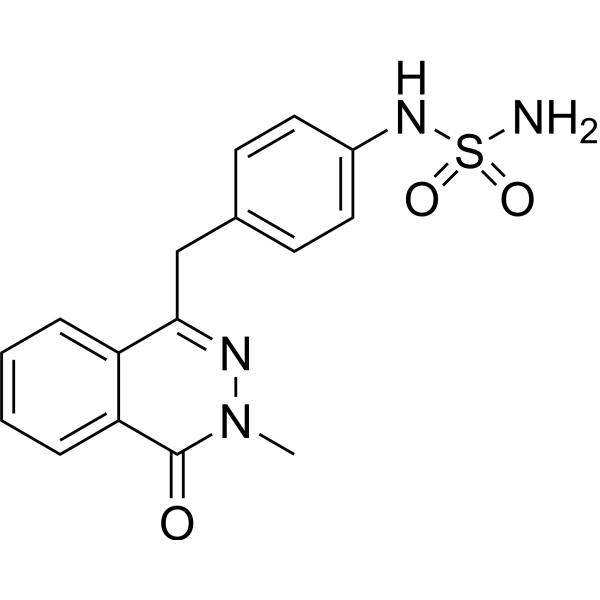
-
GC74032
Enpp-1-IN-20
Enpp-1-IN-20 (Compound 31) is an ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) inhibitor, with an IC50 of 0.09 nM.
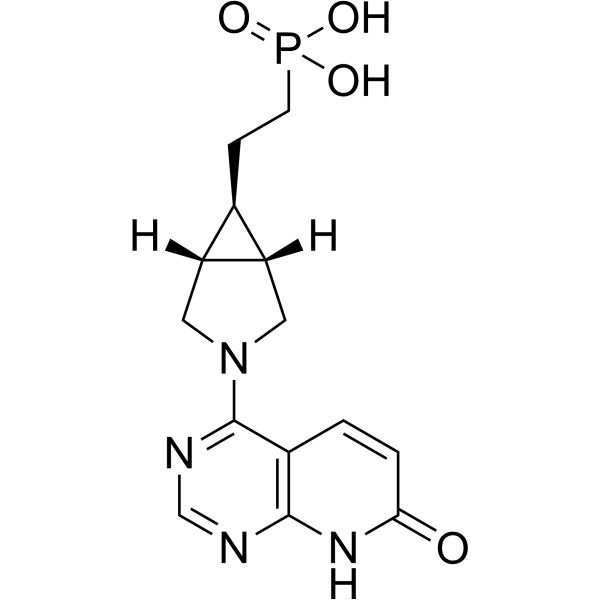
-
GC52334
ENPP1 Inhibitor 4e
cGAMP-compound 4e
An ENPP1 inhibitor
-
GC64257
Gelsevirine
Gelsevirine is the major alkaloid in Gelsemium elegans with potent anxiolytic effects.

-
GC34610
H-151
H-151 is a low MW antagonist of STING, which blocks the activation-induced palmitoylation of STING, and exhibits significant inhibitory effects on STING signalling H-151 can be used in the study of autoinflammatory diseases.

-
GC63012
IACS-8803 diammonium
IACS-8803 diammonium is a highly potent cyclic dinucleotide STING agonist. IACS-8803 diammonium has a robust systemic antitumor efficacy.

-
GC62586
IACS-8803 disodium
IACS-8803 disodium is a highly potent cyclic dinucleotide STING agonist. IACS-8803 disodium has a robust systemic antitumor efficacy.

-
GC43894
IKK2 Inhibitor VI
5-Phenyl-2-ureidothiophene-3-carboxylic Acid Amide
Inhibitor of NF-κB kinase 2 (IKK2, also known as IKKβ) acts as part of an IKK complex in the canonical NF-κB pathway, phosphorylating inhibitors of NF-κB (IκBs) to initiate signaling.
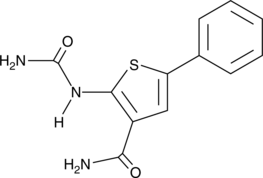
-
GC52291
KAS 08
A STING activator

-
GC91789
LB244
LB244 is an irreversible STING antagonist.
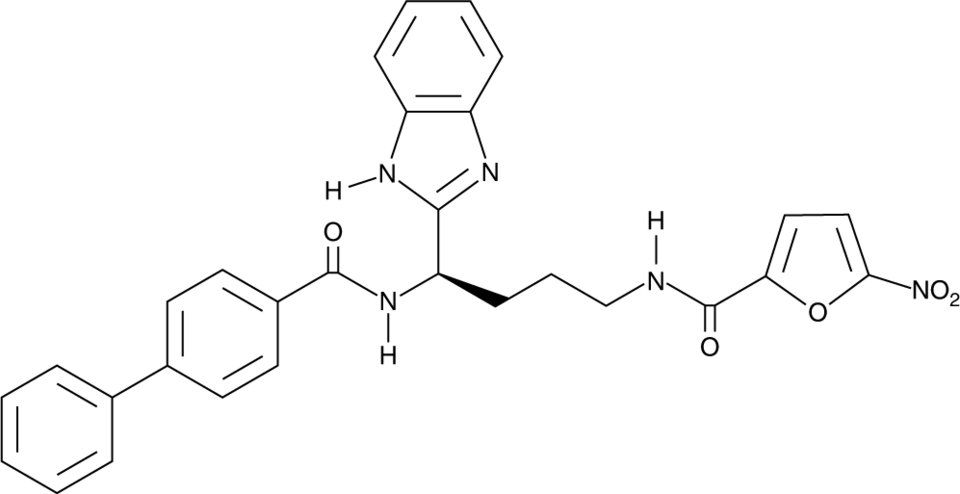
-
GC49673
M04
A STING agonist

-
GC18988
ML RR-S2 CDA (ammonium salt)
STING Inducer-1
ML RR-S2 CDA (ammonium salt) (MIW815 ammonium salt), an activator of stimulator of interferon genes (STING), leads to potent and systemic tumor regression and immunity.
-
GC61092
MSA-2
5,6-dimethoxy-γ-oxo-benzobthiophene-2-Butanoic Acid
MSA-2 is an orally available non-nucleotide interferon gene stimulator (STING) agonist, with EC50 values of 8.3μM and 24μM for human STING subtypes WT and HAQ, respectively.
-
GC63082
MSA-2 dimer
MSA-2 dimer is a selective, orally active non-nucleotide STING agonist (Kd=145 μM) with long-term antitumor and immunogenic activity.

-
GC44388
NF-κB Control
SN50M
NF-κB inhibitor is a synthetic peptide corresponding to the nuclear localization sequence (NLS) of NF-κB p105 subunit (also known as p50) appended to a hydrophobic sequence to facilitate import into living cells.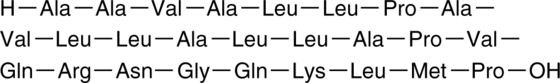
-
GC48985
NF-κB Inhibitor (trifluoroacetate salt)
SN50 Peptide
A cell-permeable peptide that blocks nuclear import of NF-κB
-
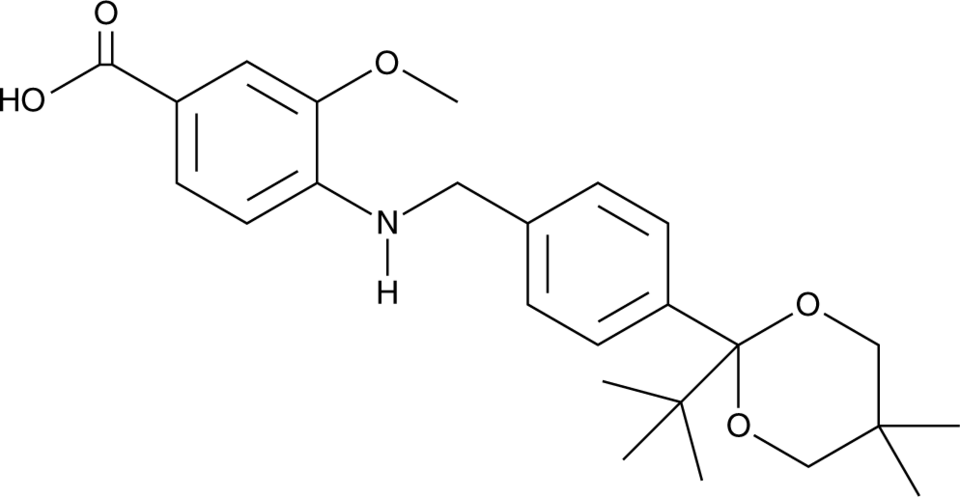
GC91777
NVS-STG2
NVS-STG2 is an agonist of stimulator of interferon genes (STING).
-
GC13693
Omaveloxolone (RTA-408)
Omaveloxolone
Omaveloxolone (RTA-408) (RTA 408) is an antioxidant inflammation modulator (AIM), which activates Nrf2 and suppresses nitric oxide (NO).
-
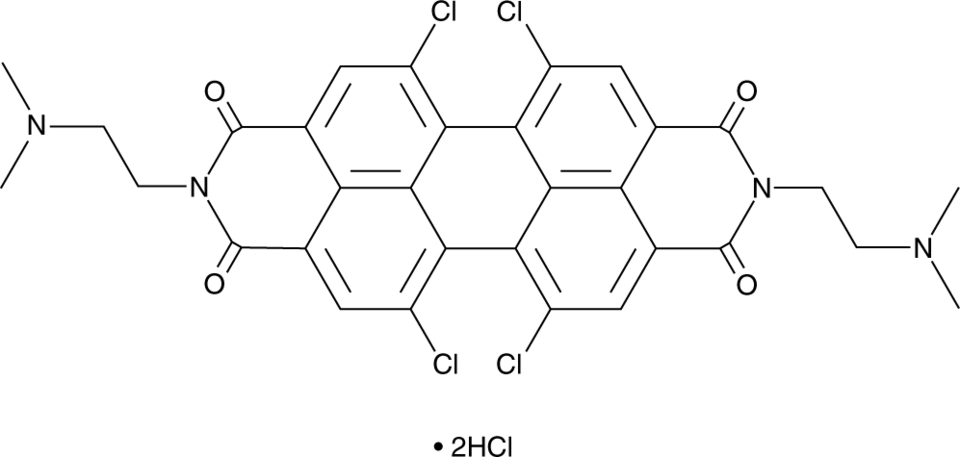
GC91595
PDIC-NC
PDIC-NC is an activator of stimulator of interferon genes (STING) and an anticancer agent.
-
GC91688
PDIC-NN
PDIC-NN is an intermediate in the synthesis of PDIC-NS , an activator of stimulator of interferon genes (STING) with anticancer activity.

-
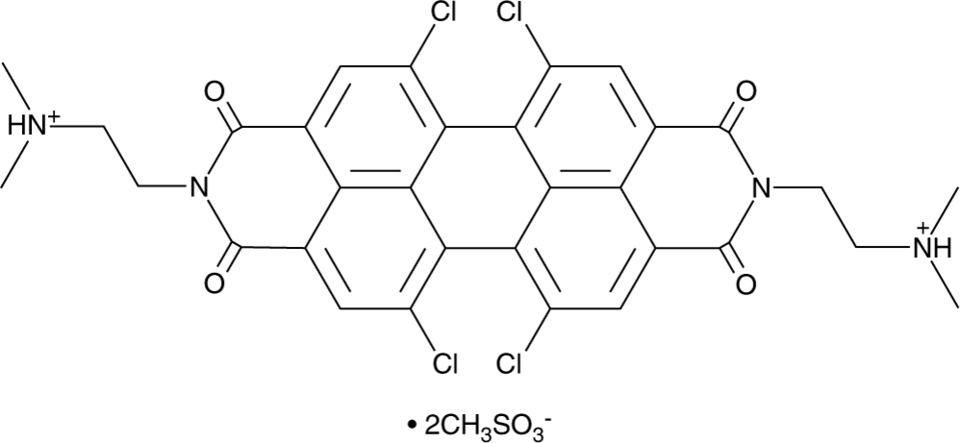
GC91820
PDIC-NS
PDIC-NS is an activator of stimulator of interferon genes (STING).
-
GC66392
PROTAC STING Degrader-1
PROTAC STING Degrader-1 (Compound SP23) is a STING PROTAC degrader with a DC50 of 3.2 μM. PROTAC STING Degrader-1 shwos anti-inflammatory activity.

-
GC91096
S-72
An inhibitor of microtubule polymerization

-
GC67719
SN-001

-
GC63905
SN-008
SN-008, a less active SN-011 analog, can be used as a negative control.

-
GC49400
SN-011
GUN35901
A STING antagonist
-
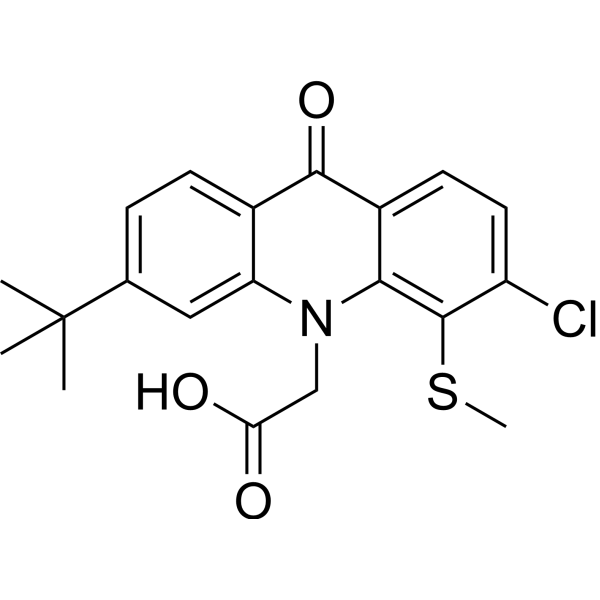
GC73865
SNX281
SNX281 is a systemically active STING agonist that binds to the STING protein, promotes signal transduction of the cGAS-STING pathway, and increases cellular responses to tumor cells.
-
GC61293
SR-717
SR-717 is a stable cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) mimetic, Antitumor activity.

-
GC44948
StA-IFN-1
StA-IFN-1 is an inhibitor of the interferon (IFN) induction pathway with an IC50 value of 4.1 μM in a GFP reporter assay for IFN induction similar to TPCA-1, which specifically inhibits the IKKβ component of the IFN induction pathway.

-
GC48109
STING Agonist 12b
A STING agonist

-
GC90784
STING Agonist 12L
A STING agonist
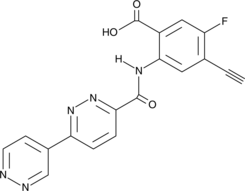
-
GC48110
STING Agonist 1a
STING Agonist 1a (1a) is a specific stimulator of interferon genes (STING) agonist.

-
GC90785
STING Agonist 23
A STING agonist
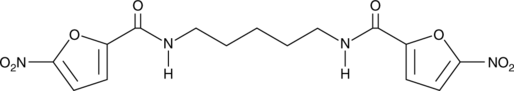
-
GC45570
STING Agonist C11

-
GC90982
STING Agonist D61
A STING agonist
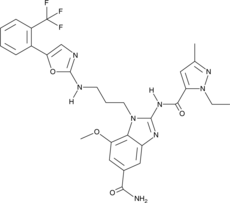
-
GC12943
STING agonist-1
STING Agonist-1
An indirect activator of STING signaling
-
GC65282
STING agonist-12
STING Agonist-12
STING agonist-12 (Compound 53) is a potent, orally active human STING activator with an EC50 of 185 nM.
-
GC73098
STING agonist-17
STING agonist-17 (compound 4a) is a potent STING agonist with an IC50 value of 0.062 nM.
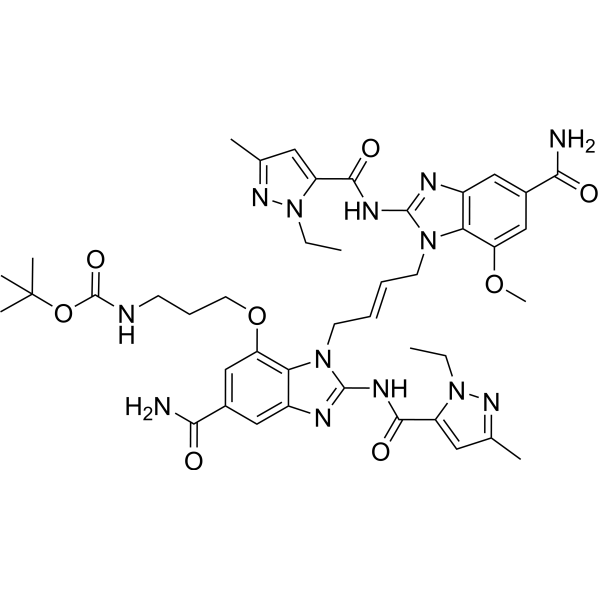
-
GC69959
STING agonist-20
STING agonist-20 (compound 95) is an effective STING agonist used in the synthesis of XMT-2056. It can be used as a vaccine adjuvant for cancer, inflammation, and research on immune diseases.

-
GC69961
STING agonist-22
STING agonist-22 (CF501) is an effective non-nucleotide STING agonist.

-
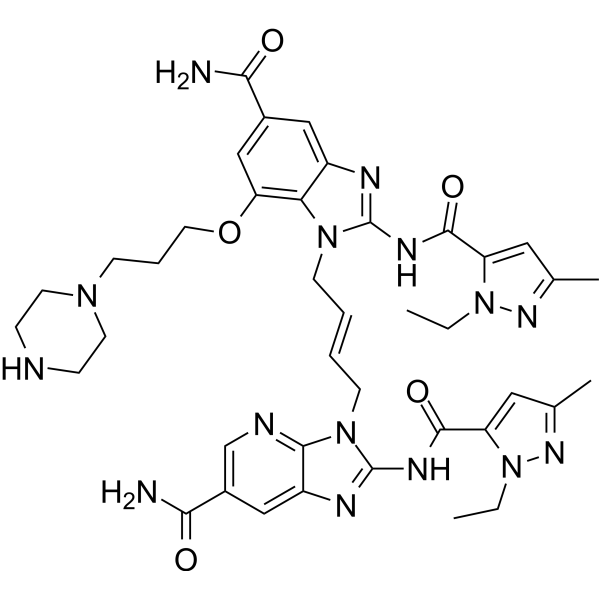
GC73407
STING agonist-26
STING agonist-26 (CF508) is a non-nucleotide small-molecule STING agonist.
-
GC37692
STING agonist-3
CS-0029879, Diamidobenzimidazole STING Agonist, diABZI-3, EX-A3217, HY-103665, STING Agonist-3
STING agonist-3, extracted from patent WO2017175147A1 (example 10), is a selective and non-nucleotide small-moleculeSTINGagonist with a pEC50 and pIC50 of 7.5 and 9.5, respectively. STING agonist-3 has durable anti-tumor effect and tremendous potential to improve treatment of cancer.
-
GC69962
STING agonist-3 trihydrochloride
STING agonist-3 trihydrochloride is derived from patent WO2017175147A1 (Example 10) and is a selective and non-nucleotide small molecule STING agonist with pEC50 and pIC50 values of 7.5 and 9.5, respectively. It has long-lasting anti-tumor effects and holds great potential in improving cancer research.

-
GC37693
STING agonist-4
CS-0087594, Diamidobenzimidazole STING Agonist-2, diABZI-4, HY-123943, STING Agonist-4, STING Agonist diABZI Compound 2
A STING agonist
-
GC66008
STING agonist-7
STING agonist-7 is a non-nucleotide STING agonist. STING agonist-7 binds selectively to mouse STING but not human STING. STING agonist-7 penetrates cell membrane poorly.




