MAPK Signaling

Targets for MAPK Signaling
- ERK(78)
- MEK1/2(63)
- NKCC(5)
- MNK(7)
- PKA(53)
- p38(98)
- Rac(2)
- Raf(63)
- RasGAP (Ras- P21)(0)
- JNK(57)
- cAMP(39)
- Protein Kinase G(2)
- RSK(30)
- Other(759)
- MAPKAPK(1)
- MKK(2)
- KLF(3)
- MAP3K(21)
- MAP4K(20)
- MAPKAPK2 (MK2)(9)
- Mixed Lineage Kinase(9)
Products for MAPK Signaling
- Cat.No. Product Name Information
-
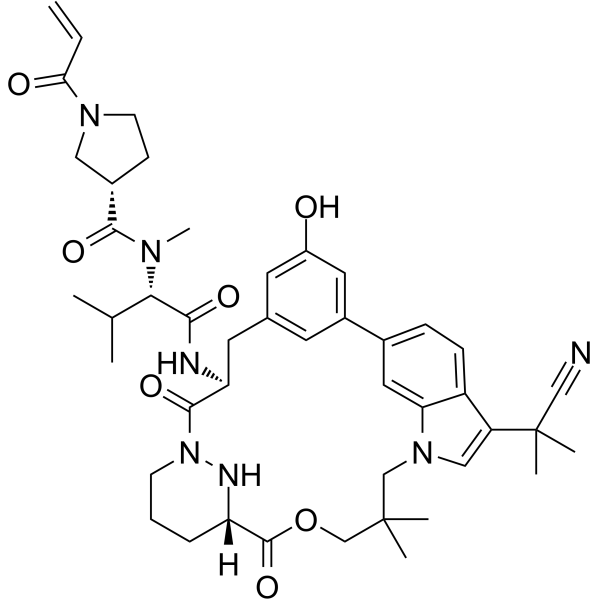
GC62504
RAS/RAS-RAF-IN-1
RAS/RAS-RAF-IN-1 is a potent RAS and RAS-RAF inhibitor. RAS/RAS-RAF-IN-1 has a KD of 5.0 μΜ-15 μΜ for cyclophilin A (CYPA) binding affinity. RAS/RAS-RAF-IN-1 has antitumor activity.
-
GC37071
Ravoxertinib hydrochloride
GDC-0994 hydrochloride
Ravoxertinib hydrochloride (GDC-0994 hydrochloride) is an orally bioavailable inhibitor selective for ERK kinase activity with IC50 of 6.1 nM and 3.1 nM for ERK1 and ERK2, respectively.
-
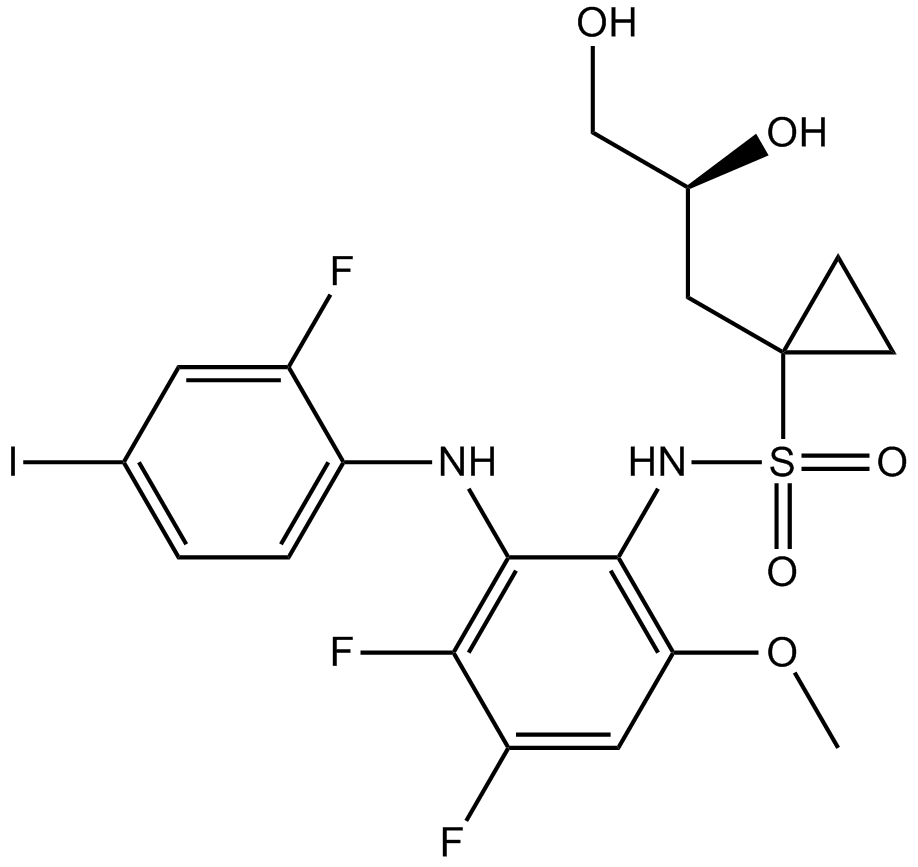
GC16872
Refametinib
BAY 869766; RDEA119;
MEK 1/ MEK 2 inhibitor
-
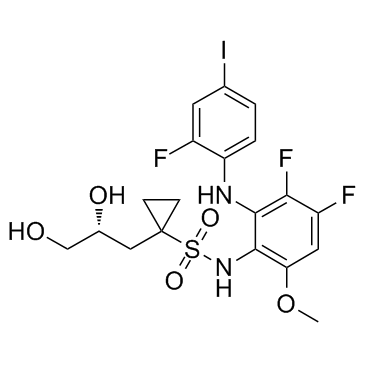
GC37516
Refametinib R enantiomer
Refametinib R enantiomer is a MEK inhibitor extracted from patent WO2007014011A2, compound 1022, has an EC50 of 2.0-15 nM.
-
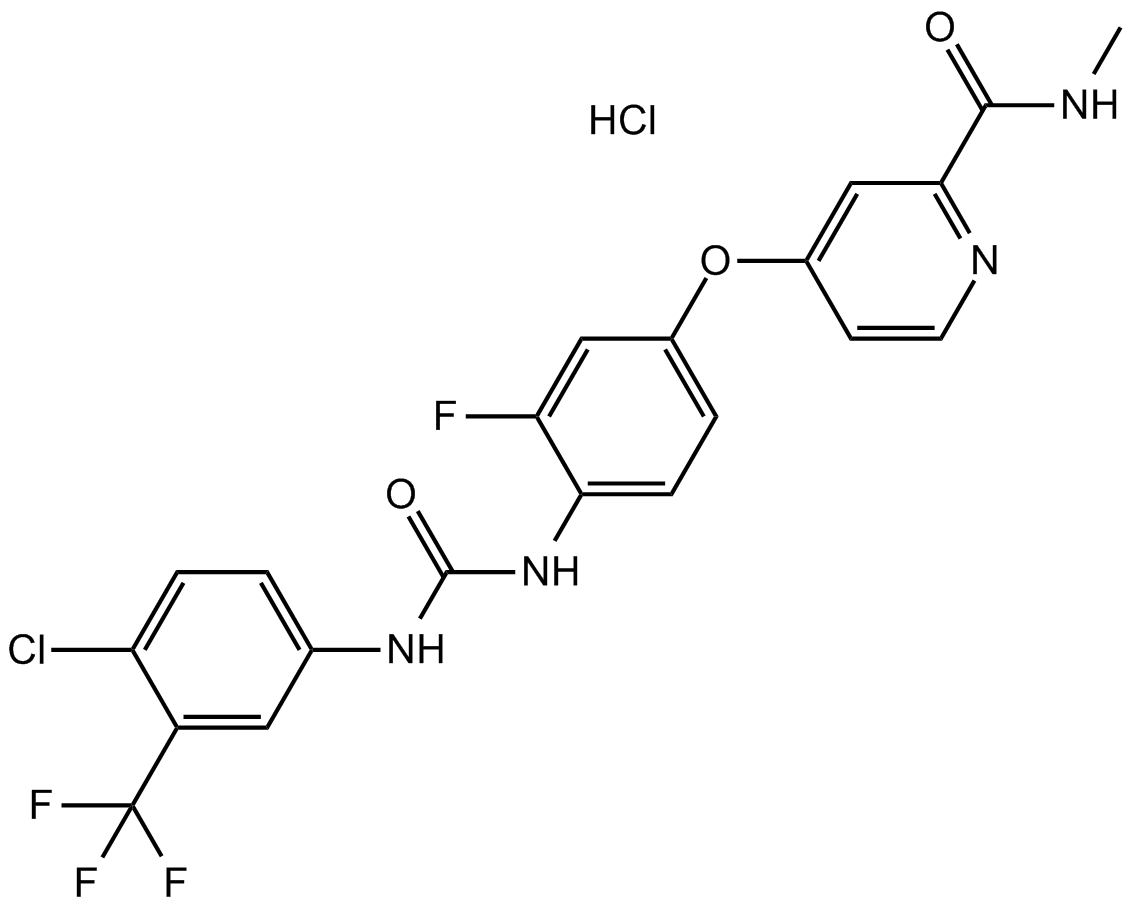
GC14606
Regorafenib hydrochloride
A multi-kinase inhibitor
-
GC14534
Regorafenib monohydrate
A multi-kinase inhibitor

-
GC40213
Regorafenib-13C-d3
Regorafenib-13C-d3 is intended for use as an internal standard for the quantification of regorafenib by GC- or LC-MS.

-
GC18751
Reticulol
6,8-dihydroxy-7-methoxy-3-methyl Isocoumarin, NSC 294978
Reticulol is an isocoumarin derivative produced by certain species of Streptomyces that inhibits cAMP phosphodiesterase (IC50 = 41 uM).
-
GC37522
RGB-286638
A multi-kinase inhibitor

-
GC37523
RGB-286638 free base
A multi-kinase inhibitor
-
GN10784
Rhoifolin
Apigenin 7-O-Neohesperidoside

-
GC50502
RI-STAD 2
AKAP disruptor; selectively binds PKA-RI with high affinity and blocks its interaction with AKAP; cell permeable

-
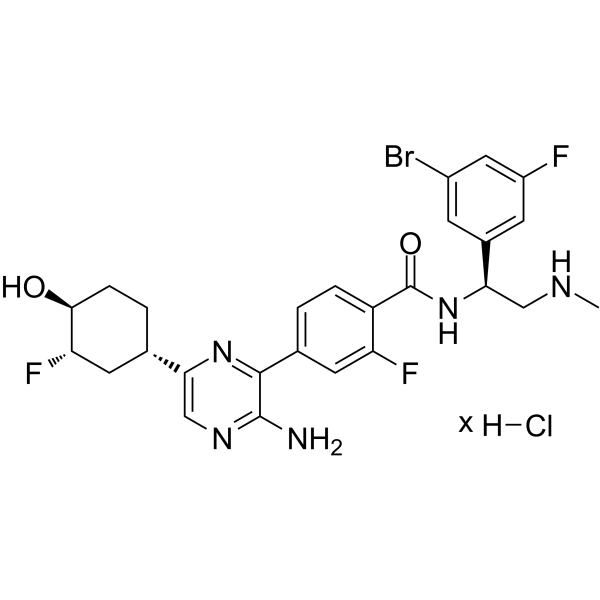
GC62448
Rineterkib hydrochloride
Rineterkib hydrochloride (compound B) is an orally available ERK1 and ERK2 inhibitor in the treatment of a proliferative disease characterized by activating mutations in the MAPK pathway. The activity is particularly related to the treatment of KRAS-mutant NSCLC, BRAF-mutant NSCLC, KRAS-mutant pancreatic cancer, KRAS-mutant colorectal cancer (CRC) and KRAS-mutant ovarian cancer. Rineterkib hydrochloride can also inhibit RAF.
-
GC69821
RLX-33
RLX-33 is an effective and selective relaxin family peptide 3 (RXFP3) antagonist with blood-brain barrier permeability. It also blocks the phosphorylation of ERK1/2 induced by relaxin 3, with IC50 values for RXFP3, ERK1, and ERK2 phosphorylation being 2.36 μM, 7.82 μM, and 13.86 μM respectively. RLX-33 can block the increase in food intake induced by the RXFP3 selective agonist R3/I5 in rats. RLX-33 can be used for research on metabolic syndrome.

-
GC50294
RMM 46
MSK/RSK family kinase inhibitor

-
GC12141
RO4987655
CH4987655
MEK inhibitor
-
GC12406
RO5126766(CH5126766)
CH5126766, VS-6766
RO5126766(CH5126766) (CH5126766) is a first-in-class dual MEK/RAF inhibitor that allosterically inhibits BRAFV600E, CRAF, MEK, and BRAF (IC50: 8.2, 56, 160 nM, and 190 nM, respectively).
-
GC38609
Rotundic acid
Rotundic acid, a triterpenoid obtained from I.

-
GC15611
Rp-8-Br-PET-cGMPS
Rp-8-bromo-PET-cGMPS
cGMP-dependent protein kinase (PKG) inhibitor
-
GC44852
Rp-8-bromo-Cyclic AMPS (sodium salt)
8-Bromoadenosine 3',5'-cyclic monophosphorothioate, Rp-isomer, Rp-8-BrcAMPS, Rp-8-bromocAMPS
Rp-8-bromo-Cyclic AMPS (Rp-8-bromo-cAMPS) is a cell-permeable cAMP analog that combines an exocyclic sulfur substitution in the equatorial position of the cyclophosphate ring with a bromine substitution in the adenine base of cAMP.
-
GC44853
Rp-8-CPT-Cyclic AMP (sodium salt)
Rp-8-CPT-cAMP
Rp-8-CPT-Cyclic AMP (sodium salt), a cAMP analog, is a potent and competitive antagonist of cAMP-induced activation of cAMP-dependent PKA I and II.
-
GC63176
Rp-cAMPS sodium salt
Rp-cAMPS sodium salt, a cAMP analog, is a potent, competitive cAMP-induced activation of cAMP-dependent PKA I and II (Kis of 12.5 ?M and 4.5 ?M, respectively) antagonist.

-
GC62269
RRD-251
RRD-251 is an inhibitor of retinoblastoma tumor suppressor protein (Rb)-Raf-1 interaction, with potent anti-proliferative, anti-angiogenic and anti-tumor activities.
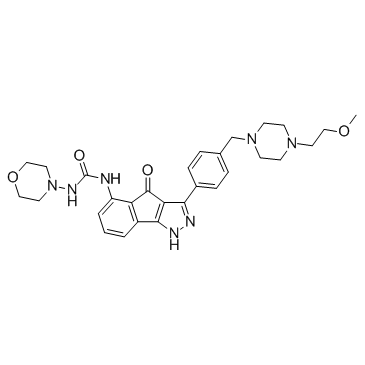
-
GC13142
RWJ 67657
JNJ-3026582
orally active inhibitor of the MAP kinases p38α and p38β
-
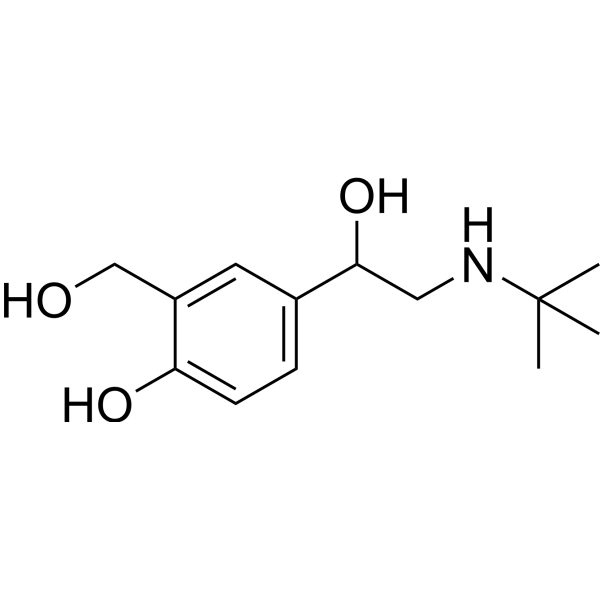
GC71682
Salbutamol
Salbutamol (Albuterol) is a short-acting beta-2 adrenergic receptor agonist with oral activity.
-
GC18273
SB 202190 (hydrochloride)
SB 202190 is a potent, selective, and cell-permeable inhibitor of p38 MAP kinases, inhibiting p38α (SAPK2A, MAPK14) and p38β (SAPK2B, MAPK11) with IC50 values of 50 and 100 nM, respectively.

-
GC16019
SB 202474
a negative control in studies of p38 inhibition

-
GC13001
SB 203580 hydrochloride
PB 203580, RWJ 64809
Adezmapimod (SB 203580) hydrochloride is a selective and ATP-competitive p38 MAPK inhibitor with IC50s of 50 nM and 500 nM for SAPK2a/p38 and SAPK2b/p38β2, respectively.
-
GC10054
SB 239063
A selective p38 MAPK inhibitor

-
GC37595
SB 242235
SB-242235 is a potent and selective p38 MAP kinase inhibitor, with an IC50 of 1.0 μM in primary human chondrocytes.

-
GC11922
SB 706504
p38 MAPK inhibitor

-
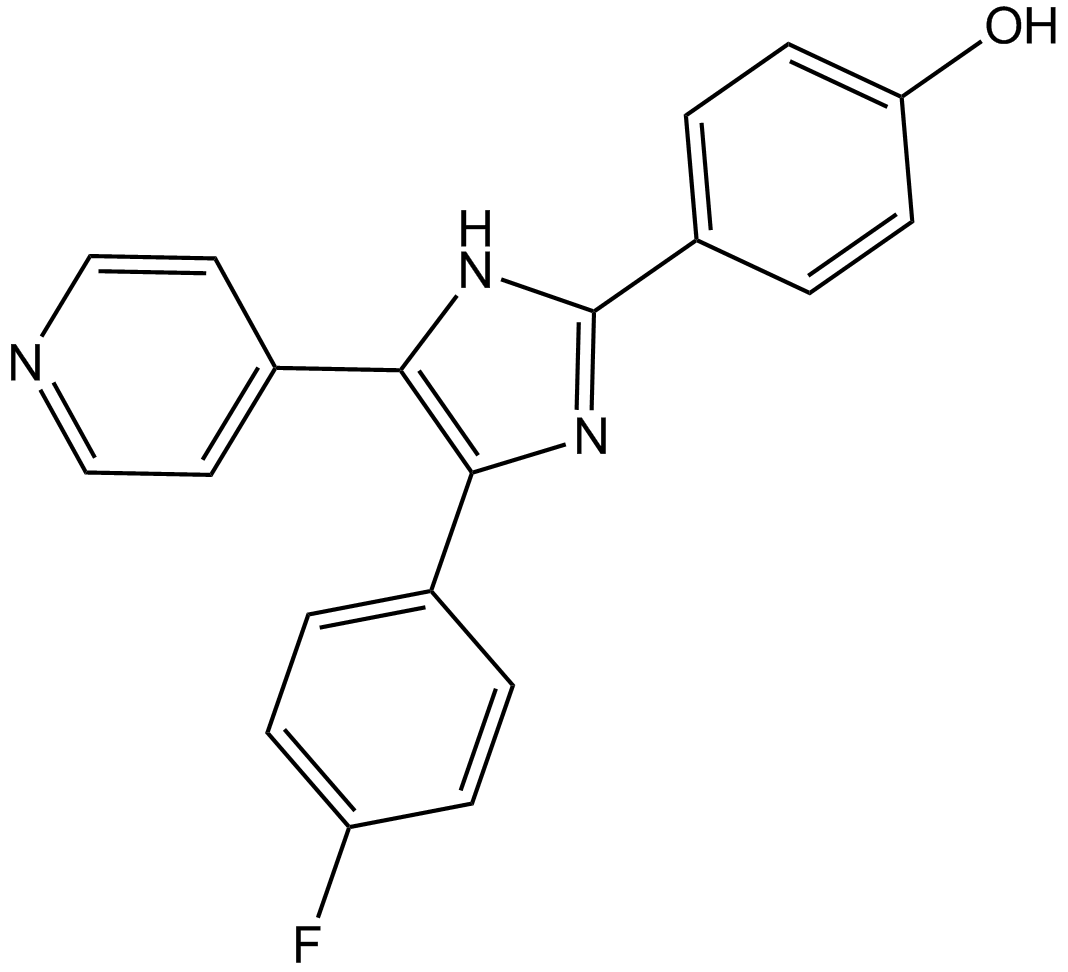
GC13968
SB202190 (FHPI)
SB202190 (FHPI) is a selective p38 MAP kinase inhibitor with IC50s of 50 nM and 100 nM for p38α and p38β2, respectively.
-
GC11890
SB590885
Potent B-Raf inhibitor

-
GC16001
SCH772984
SCH772984 is a novel, potent, ATP-competitive inhibitor of ERK1 and ERK2 with IC50 values of 4nM and 1nM, respectively.

-
GC14647
SCH772984 HCl
ERK1/2 inhibitor

-
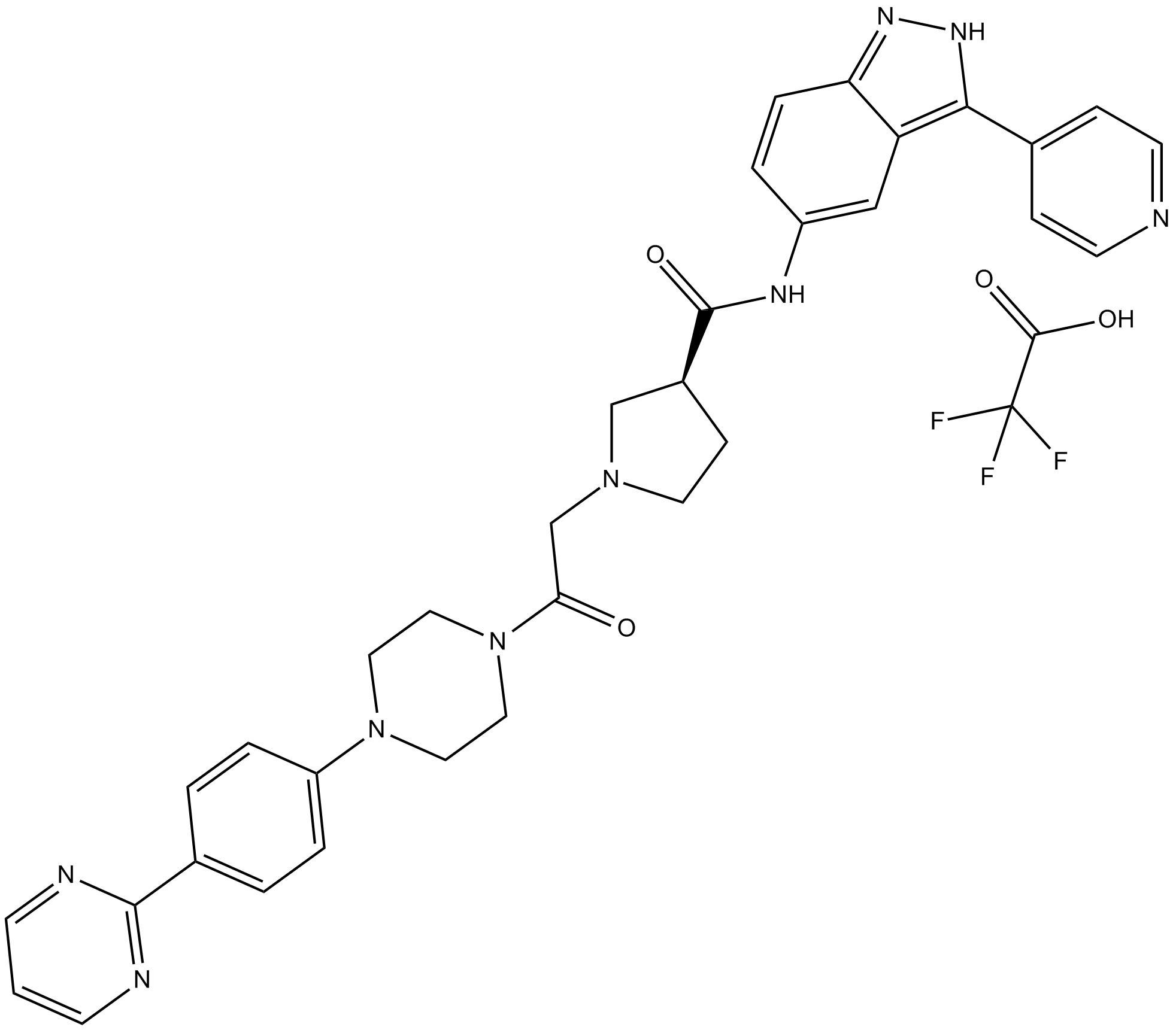
GC15044
SCH772984 TFA
ERK1/2 inhibitor
-
GC19325
SD 0006
SD-006
SD-06 is a p38 MAP kinase inhibitor; inhibits p38α with an IC50 value of 170 nM and inhibits LPS-stimulated TNF-release in rats (83% inhibition at 1mg/kg, po).IC50 value: 170 nM.
-
GC14982
SD 169
5-Carbamoylindole
selective ATP competitive inhibitor of the MAP kinases p38α and p38β
-
GC12124
Selonsertib (GS-4997)
GS-4977, GS-4997
Selonsertib (GS-4997) (GS-4997), an orally bioavailable, selective apoptosis signal-regulating kinase 1 (ASK1) inhibitor with a pIC50 of 8.3, has been evaluated as an experimental treatment for diabetic nephropathy and kidney fibrosis.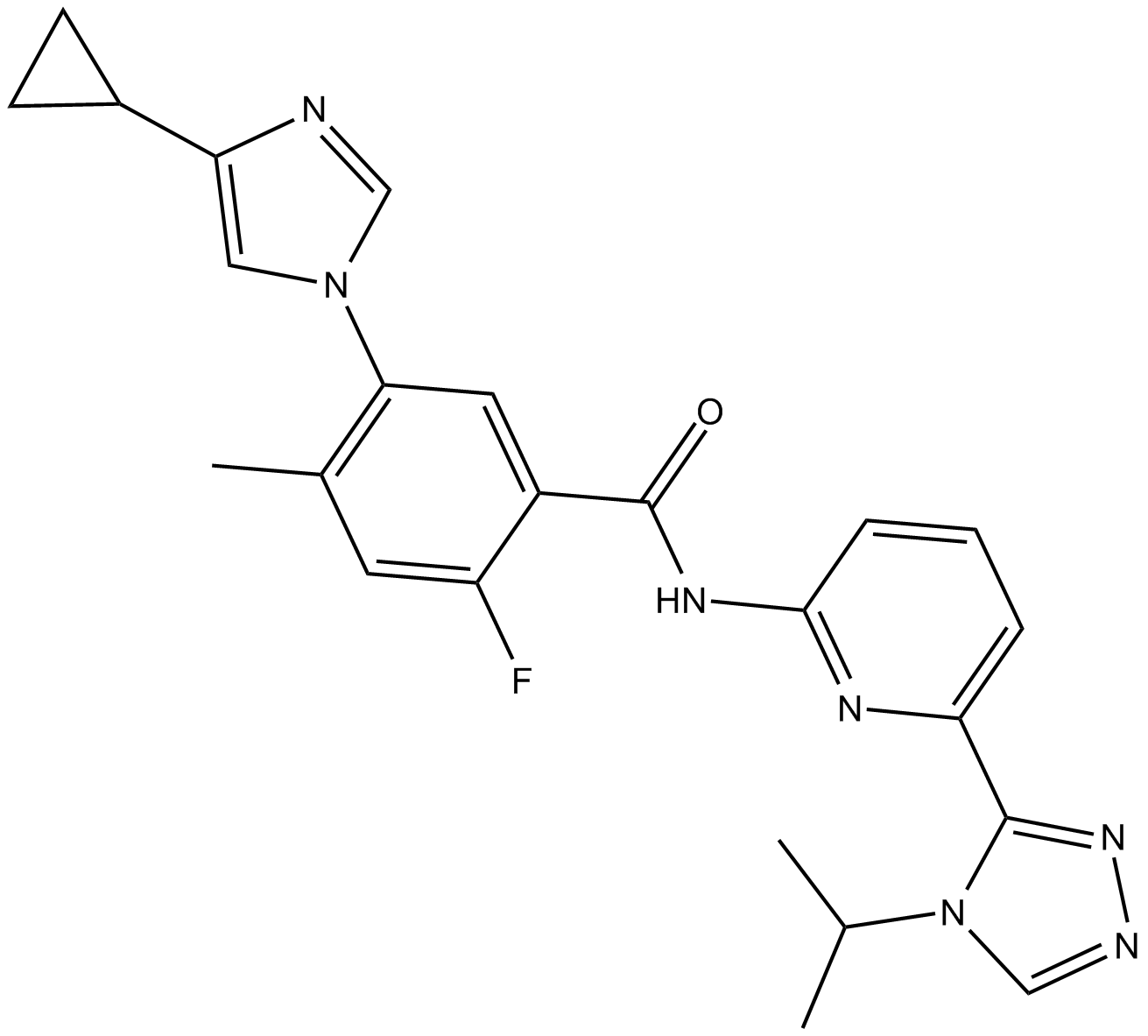
-
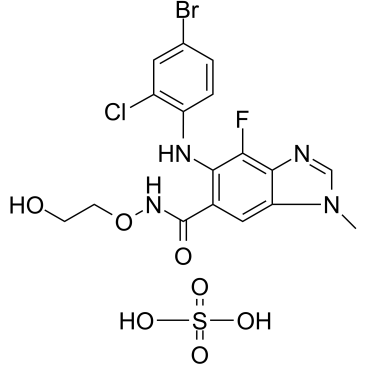
GC38653
Selumetinib sulfate
Selumetinib (AZD6244) is selective, non-ATP-competitive oral?MEK1/2?inhibitor, with an IC50 of 14 nM for MEK1. Selumetinib (AZD6244) inhibits ERK1/2 phosphorylation.
-
GC39815
Semapimod tetrahydrochloride
CNI-1493; CPSI-2364 tetrahydrochloride
Semapimod tetrahydrochloride (CNI-1493), an inhibitor of proinflammatory cytokine production, can inhibit TNF-α, IL-1β, and IL-6.
-
GC10005
SEP-0372814
A PDE10A inhibitor

-
GC33229
SJFα
SJFα is a 13-atom linker PROTAC based on von Hippel-Lindau ligand. SJFα degrades p38α with a DC50 of 7.16nM, but is far less effective at degrading p38δ (DC50=299nM) and does not degrade the other p38 isoforms (β and γ) at concentrations up to 2.5μM.

-
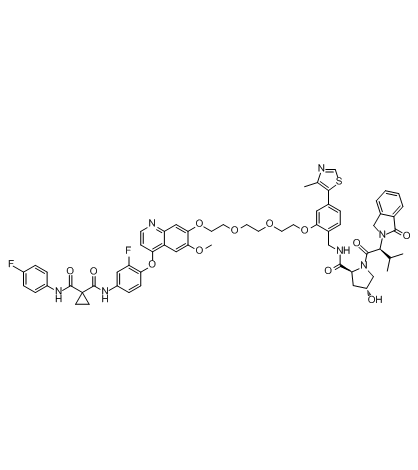
GC33238
SJFδ
SJFδ is a 10-atom linker PROTAC based on von Hippel-Lindau ligand. SJFδ degrades p38δ with a DC50 of 46.17nM, but does not degrade p38α, p38β, or p38γ.
-
GC30646
Skatole(3-Methylindole)
3-Methyl-1H-indole
Skatole(3-Methylindole) is produced by intestinal bacteria, regulates intestinal epithelial cellular functions through activating aryl hydrocarbon receptors and p38.
-
GC13578
Skepinone-L
Skepinone L
An inhibitor of p38 MAPK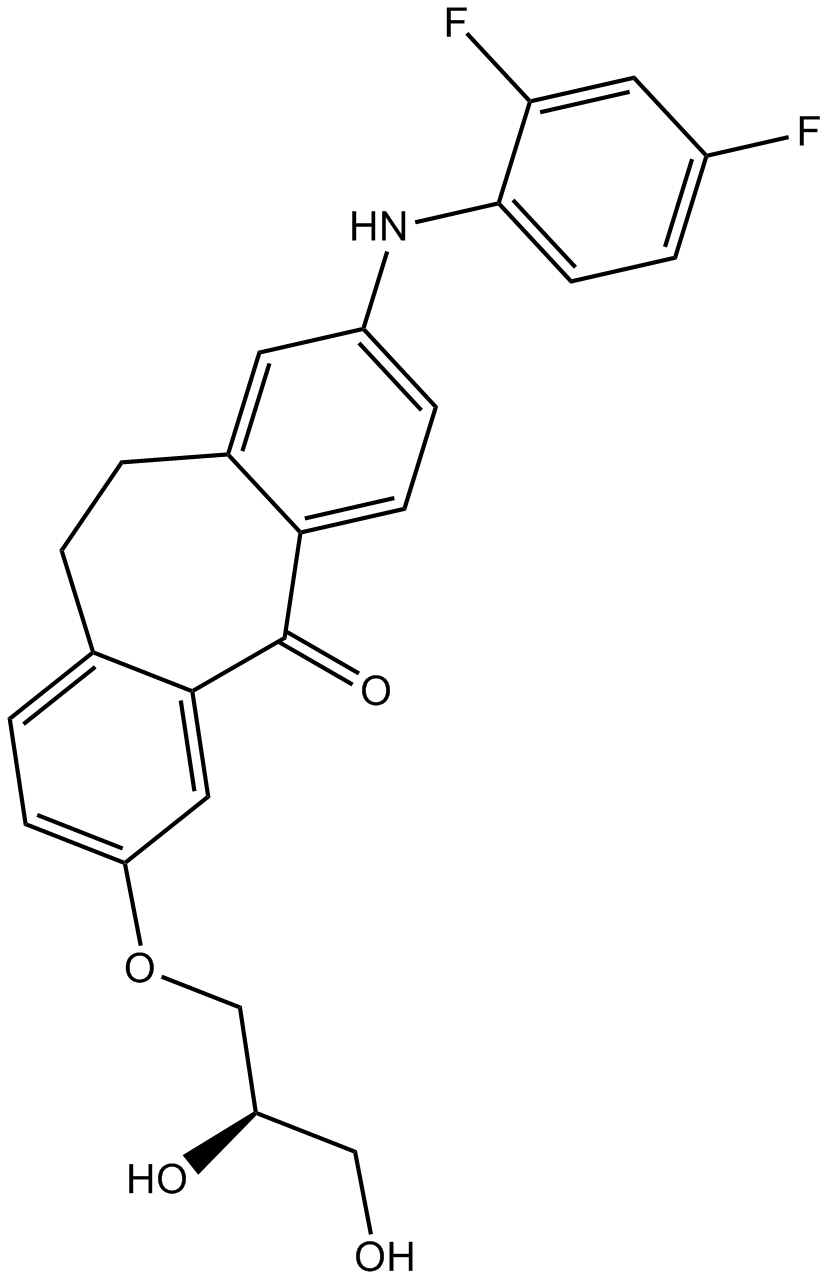
-
GC17725
SKF 86002 dihydrochloride
p38 MAP kinase inhibitor

-
GC37646
SKF-86002
SKF-86002 is an orally active p38 MAPK inhibitor, with anti-inflammatory, anti-arthritic and analgesic activities.

-
GC18532
Skyrin
Endothianin, Rhodophyscin
Skyrin is a fungal metabolite characterized by a bisanthraquinone structure.
-
GC16313
SL 0101-1
ribosomal S6 kinase inhibitor

-
GC15359
SL-327
Selective MEK1/2 inhibitor

-
GC32937
SLV-2436 (SEL201-88)
SEL201-88; SEL-201
SLV-2436 (SEL201-88) is a highly potent and ATP-competitive inhibitor of MNK1 and MNK2 with IC50s of 10.8 nM and 5.4 nM, respectively.
-
GC62675
SM1-71
SM1-71 (compound 5) is a potent TAK1 inhibitor, with a Ki of 160 nM, it also can covalently inhibit MKNK2, MAP2K1/2/3/4/6/7, GAK, AAK1, BMP2K, MAP3K7, MAPKAPK5, GSK3A/B, MAPK1/3, SRC, YES1, FGFR1, ZAK (MLTK), MAP3K1, LIMK1 and RSK2. SM1-71 can inhibit proliferation of multiple cancer cell lines.

-
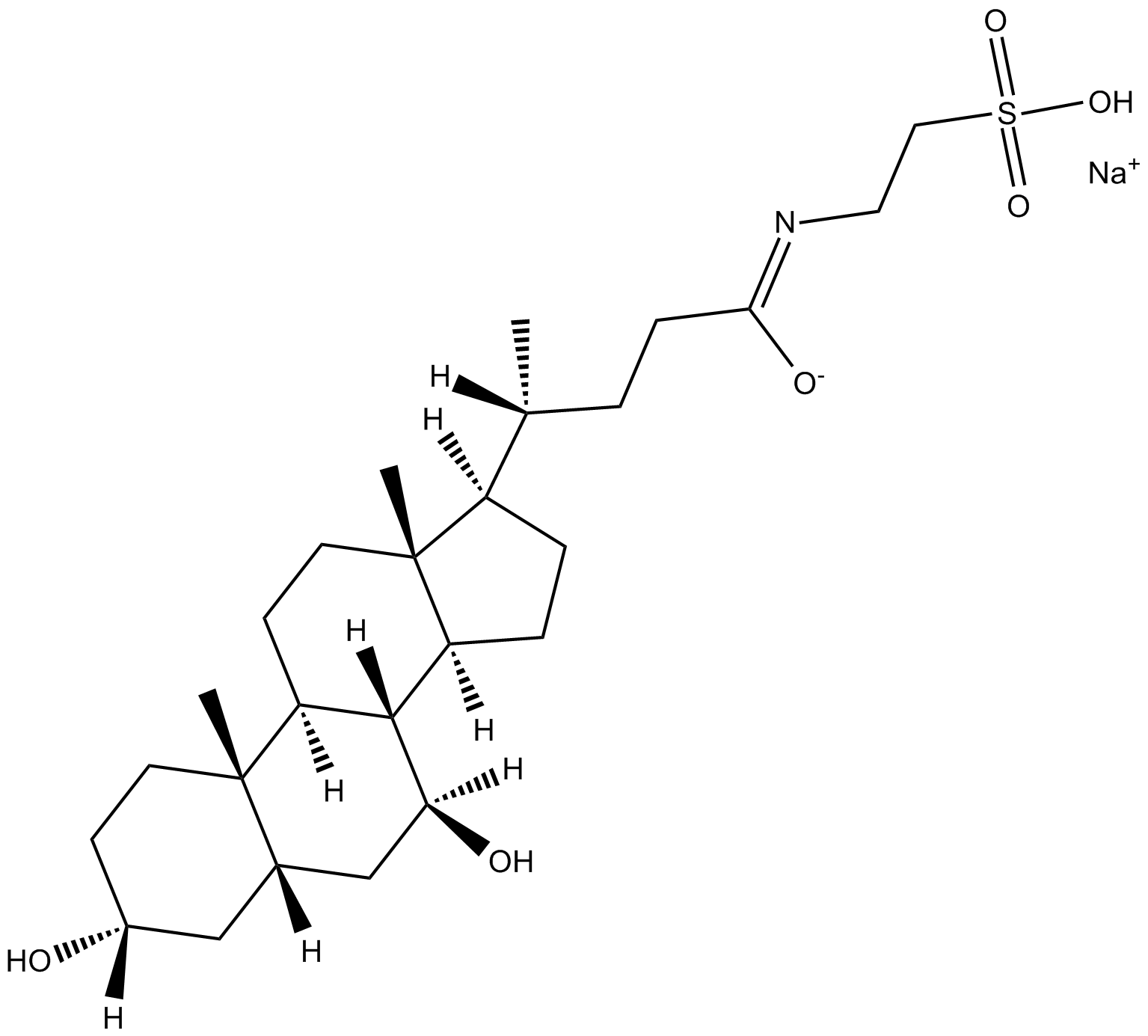
GC17425
Sodium Tauroursodeoxycholate (TUDC)
Tauroursodeoxycholate (Tauroursodeoxycholic acid; TUDCA) sodium is an endoplasmic reticulum (ER) stress inhibitor. Tauroursodeoxycholate significantly reduces expression of apoptosis molecules, such as caspase-3 and caspase-12. Tauroursodeoxycholate also inhibits ERK.
-
GC17369
Sorafenib
BAY 43-9006
Sorafenib acts as a multi-kinase inhibitor, targeting Raf-1 and B-Raf with IC50 values of 6 nM and 22 nM, respectively.

-
GC37664
Sorafenib (D3)
BAY 43-9006-d3
An internal standard for the quantification of sorafenib
-
GC37665
Sorafenib (D4)
Bay 43-9006-d4
Sorafenib (D4) (Bay 43-9006-d4) is the deuterium labeled Sorafenib. Sorafenib is a multikinase inhibitor IC50s of 6 nM, 20 nM, and 22 nM for Raf-1, B-Raf, and VEGFR-3, respectively.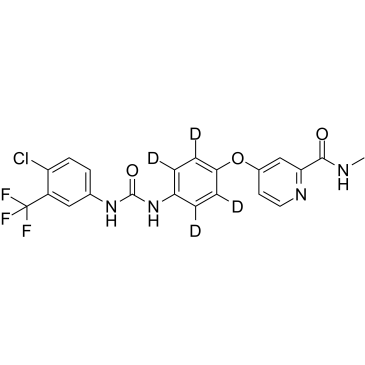
-
GC44917
Sorafenib N-oxide
BAY 67-3472
Sorafenib N-oxide is an active metabolite of sorafenib, an inhibitor of Raf-1, B-RAF, and receptor tyrosine kinases.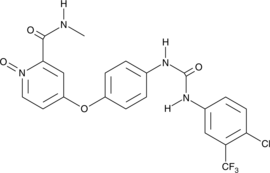
-
GC10428
Sp-5,6-dichloro-cBIMPS (sodium salt)
Sp-5,6-DCI-cBIMPS
PKA activator
-
GC44920
Sp-8-bromo-Cyclic AMPS (sodium salt)
8Bromoadenosine 3',5'cyclic Monophosphothioate SPIsomer, Sp8bromocAMPS
Sp-8-bromo-cyclic AMPS (Sp-8-bromo-cAMPS) is a cell-permeable, cAMP analog that combines an exocyclic sulfur substitution in the axial position of the cyclophosphate ring with a bromine substitution in the adenine base of cAMP.
-
GC61747
Sp-cAMPS
Sp-cAMPS, a cAMP analog, is potent activator of cAMP-dependent PKA I and PKA II.

-
GC16652
SR 3576
JNK3 inhibitor, potent and selective

-
GC30864
SR-3306
SR-3306 is a selective, potent, highly brain penetrant JNK inhibitor.

-
GC64287
SR15006
SR15006 is a inhibitor of Krüppel-like factor 5 (KLF5) with an IC50 of 41.6 nM in DLD-1/pGL4.18hKLF5p cells).

-
GC50406
st-Ht31
Inhibits PKA/AKAP interactions; cell permeable

-
GC50407
st-Ht31 P
Negative control for st-Ht31

-
GC50501
STAD 2
AKAP disruptor; selectively binds PKA-RII and blocks its interaction with AKAP; cell permeable

-
GC15299
Staurosporine(CGP 41251)
Stsp
Staurosporin, a small kinase inhibitor, is an alkaloid derived from the bacterium Streptomyces staurosporeus.
-
GC11542
SU 3327
SU 3327
JNK inhibitor
-
GC13825
TA 01
CK1ε, CK1δ,and p38α inhibitor

-
GC11635
TA 02
p38 MAPK inhibitor

-
GC16543
TAK-715
A p38 MAPK inhibitor

-
GC10209
TAK-733
MEK allosteric site inhibitor

-
GC62497
TAK1-IN-2
TAK1-IN-2 is a potent and selective TAK1 inhibitor, with an IC50> of 2 nM.

-
GC69984
TAK1-IN-4
TAK1-IN-4 (Compound 14) is a TAK1 inhibitor.

-
GC49700
Takeda-6d

-
GC32687
Takinib
EDHS-206
A TAK1 inhibitor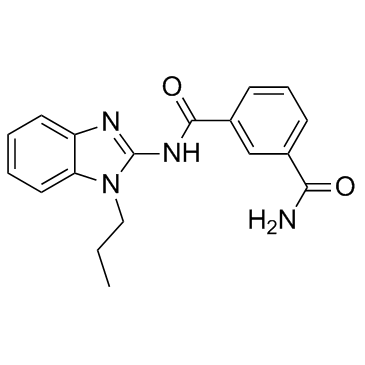
-
GC34072
Talmapimod (SCIO-469)
Talmapimod
Talmapimod (SCIO-469) (SCIO-469) is an orally active, selective, and ATP-competitive p38α inhibitor with an IC50 of 9 nM. Talmapimod (SCIO-469) shows about 10-fold selectivity over p38β, and at least 2000-fold selectivity over a panel of 20 other kinases, including other MAPKs.
-
GC25982
Tanzisertib(CC-930)
JNK-930, JNKI-1
Tanzisertib (CC-930, JNK-930, JNKI-1) is kinetically competitive with ATP in the JNK-dependent phosphorylation of the protein substrate c-Jun and potent against all isoforms of JNK (Ki(JNK1) = 44 ± 3 nM, IC50(JNK1) = 61 nM, Ki(JNK2) = 6.2 ± 0.6 nM, IC50(JNK2) = 5 nM, IC50(JNK3) = 5 nM) and selective against MAP kinases ERK1 and p38a with IC50 of 0.48 and 3.4 μM respectively.
-
GC34181
Tauroursodeoxycholate (TUDCA)
3α,7β-dihydroxy-5β-cholanoyl Taurine, TUDCA, UR-906
Tauroursodeoxycholate (TUDCA) (Tauroursodeoxycholic acid) is an endoplasmic reticulum (ER) stress inhibitor. Tauroursodeoxycholate (TUDCA) significantly reduces expression of apoptosis molecules, such as caspase-3 and caspase-12. Tauroursodeoxycholate (TUDCA) also inhibits ERK.
-
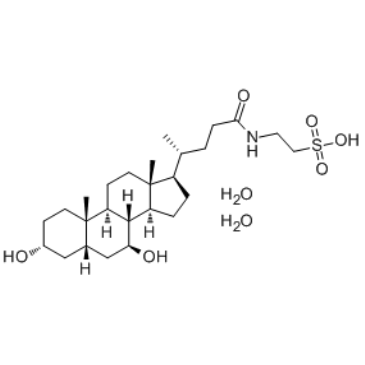
GC34831
Tauroursodeoxycholate dihydrate
Tauroursodeoxycholate (Tauroursodeoxycholic acid; TDUCA) dihydrate is an endoplasmic reticulum (ER) stress inhibitor. Tauroursodeoxycholate significantly reduces expression of apoptosis molecules, such as caspase-3 and caspase-12. Tauroursodeoxycholate also inhibits ERK.
-
GC34057
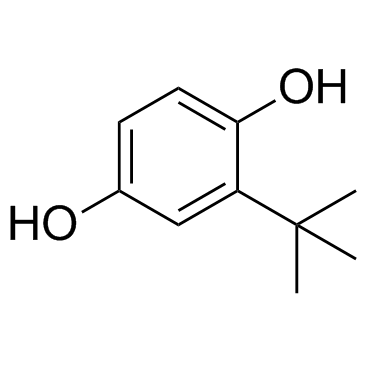
TBHQ (tert-Butylhydroquinone)
TBHQ(tert-Butylhydroquinone) is a powerful phenolic antioxidant capable of reducing oxidative stress and inflammatory reactions.
-
GC14853
TC ASK 10
ASK1 inhibitor
-
GC34140
TC13172
TC13172 is a mixed lineage kinase domain-like protein (MLKL) inhibitor with an EC50 value of 2 nM for HT-29 cells.

-
GC11488
TCS JNK 5a
cJun Nterminal Kinase Inhibitor IX, TCS JNK 5a
JNK2 and JNK3 inhibitor
-
GC17282
TCS JNK 6o
JNK Inhibitor VIII
TCS JNK 6o (TCS JNK 6o) is a c-Jun N-terminal kinases (JNK-1, -2, and -3) inhibitor with Ki values of 2 nM, 4 nM, 52 nM, respectively, and has IC50 values of 45 nM and 160 nM for JNK-1 and -2, respectively.
-
GC39074
Tenuifoliside A
Tenuifoliside A is isolated from Polygala tenuifolia, has anti-apoptotic and antidepressant-like effects.

-
GC41573
Theaflavin 3,3'-digallate
TF3, TFDG
Theaflavin 3,3'-digallate (TF-3; ZP10) is a potent inhibitor of Zika virus (ZIKV) protease with an IC50 value of 2.3μM.
-
GC68350
Tinlorafenib

-
GC50295
TL4-12
Potent MAP4K2 (GCK) inhibitor
-
GC31647
Tomatidine
Tomatidine acts as an anti-inflammatory agent by blocking NF-κB and JNK signaling.

-
GC45064
Tomatidine (hydrochloride)
Tomatidine is a steroidal alkaloid that has been found in the skins and leaves of tomatoes.

-
GC19131
Tomivosertib
eFT508 is a potent, highly selective, and orally bioavailable MNK1 and MNK2 inhibitor, with IC50s of 1-2 nM against both isoforms.

-
GC45067
Tpl2 Kinase Inhibitor
c-Cot Kinase Inhibitor, MAP3K8 Kinase Inhibitor, Tumor Progression Locus 2 Kinase Inhibitor
Tpl2 Kinase Inhibitor (Compound 1) is a potent and selective Tpl2 (COT kinase, MAP3K8) inhibitor, plays an important role in the regulation of the inflammatory response and the progression of some cancers.
-
GC49150
Tpl2 Kinase Inhibitor (hydrochloride)
c-Cot Kinase Inhibitor, MAP3K8 Kinase Inhibitor, Tumor Progression Locus 2 Kinase Inhibitor
A Tpl2 inhibitor
-
GC70053
Trametiglue
Trametiglue is a derivative of Trametinib that binds to KSR-MEK and RAF-MEK with unprecedented potency and selectivity through a unique interface interaction.

-
GC13508
Trametinib (GSK1120212)
GSK1120212, JTP-74057
Trametinib (GSK1120212, JTP-74057) is a second-generation small molecule inhibitor of MEK kinase.
-
GC15260
Trametinib DMSO solvate

-
GC30162
trans-Zeatin
A cytokinin plant growth regulator

-
GC45099
U-0126
A MEK inhibitor



