Tyrosine Kinase
- IGFBR(1)
- Bcr-Abl(67)
- Ack1(2)
- Axl(6)
- ALK(56)
- BMX Kinase(3)
- Broad Spectrum Protein Kinase Inhibitor(10)
- c-FMS(35)
- c-Kit(56)
- c-MET(84)
- c-RET(0)
- CSF-1R(3)
- DDR1/DDR2 Receptor(1)
- EGFR(235)
- EphB4(1)
- FAK(33)
- FGFR(95)
- FLT3(85)
- HER2(12)
- IGF1R(33)
- Insulin Receptor(44)
- IRAK(28)
- ITK(10)
- Lck(1)
- LRRK2(22)
- PDGFR(104)
- PTKs/RTKs(3)
- Pyk2(6)
- ROR(38)
- Spleen Tyrosine Kinase (Syk)(32)
- Src(97)
- Tie-2 (4)
- Trk(39)
- VEGFR(181)
- Kinase(0)
- Discoidin Domain Receptor(15)
- DYRK(29)
- Ephrin Receptor(12)
- ROS(14)
- RET(29)
- TAM Receptor(29)
Products for Tyrosine Kinase
- Cat.No. Product Name Information
-
GC13424
LY2874455
LY 2874455; LY-2874455
A pan-FGFR inhibitor
-
GC40865
LYG-202
LYG-202 is a synthetic flavonoid with anticancer and anti-angiogenic activities.

-
GC69418
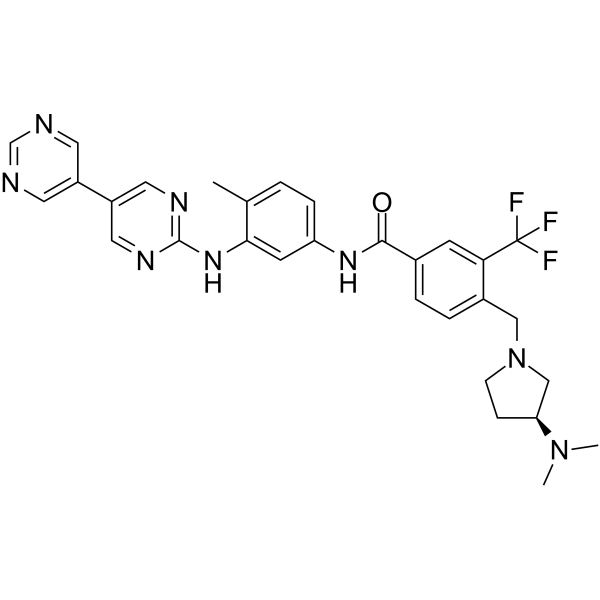
Lyn-IN-1
Bafetinib analog
Lyn-IN-1 (Bafetinib analog) is a highly active dual inhibitor of Bcr-Abl and Lyn.
-
GC62314
M4205
M4205 is a c-KIT inhibitor, with an IC50 of 10 nM for c-KIT V654A. M4205 has high activity on c-KIT mutations in exon 11, 13, 17.

-
GC68304
Margetuximab

-
GC13410
Masitinib (AB1010)
AB-1010, Masican, Masiviera
Masitinib (AB1010) (AB1010) is a potent, orally bioavailable, and selective inhibitor of c-Kit (IC50=200 nM for human recombinant c-Kit). It also inhibits PDGFRα/β (IC50s=540/800 nM), Lyn (IC50= 510 nM for LynB), Lck, and, to a lesser extent, FGFR3 and FAK. Masitinib (AB1010) (AB1010) has anti-proliferative, pro-apoptotic activity and low toxicity.
-
GC36546
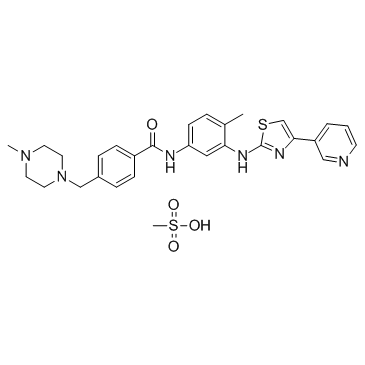
Masitinib mesylate
Masitinib mesylate (AB-1010 mesylate) is a potent, orally bioavailable, and selective inhibitor of c-Kit (IC50=200 nM for human recombinant c-Kit). It also inhibits PDGFRα/β (IC50s=540/800 nM), Lyn (IC50= 510 nM for LynB), Lck, and, to a lesser extent, FGFR3 and FAK. Masitinib mesylate (AB-1010 mesylate) has anti-proliferative, pro-apoptotic activity and low toxicity.
-
GC69436
Matuzumab
EMD 72000
Matuzumab (EMD 72000) is a humanized monoclonal antibody that can block EGFR activation and downstream signaling, inhibiting tumor growth.

-
GC66349
Mavrilimumab
CAM 3001
Mavrilimumab (CAM 3001) is a monoclonal antibody that binds to the α subunit of the granulocyte-macrophage colony stimulating factor (GM-CSF) receptor and blocks intracellular signalling downstream of GM-CSF. GM-CSF might be a mediator of the hyperactive inflammatory response associated with respiratory failure and death.
-
GC65179
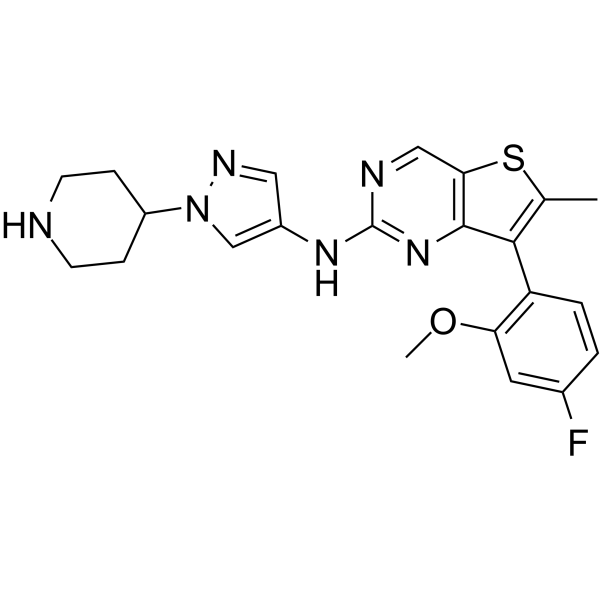
MAX-40279
MAX-40279 is a dual and potent inhibitor of FLT3 kinase and FGFR kinase.
-
GC64583
MAX-40279 hemiadipate
MAX-40279 hemiadipate is a dual and potent inhibitor of FLT3 kinase and FGFR kinase.

-
GC64582
MAX-40279 hemifumarate
MAX-40279 hemifumarate is a dual and potent inhibitor of FLT3 kinase and FGFR kinase.

-
GC16483
MAZ51
VEGFR3 antagonist

-
GC64710
MC-Val-Cit-PAB-Amide-TLR7 agonist 4
MC-Val-Cit-PAB-Amide-TLR7 agonist 4 (example 15) is a HER2-TLR7 and HER2-TLR8 immune agonist conjugate.

-
GC69460
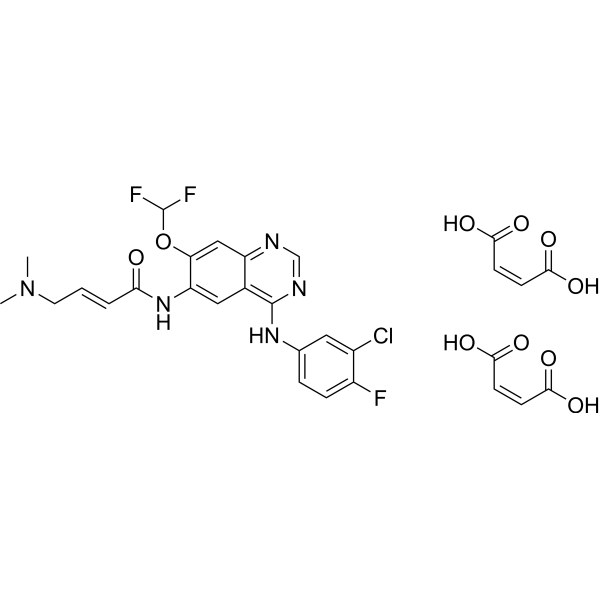
Mefatinib
Mifanertinib dimaleate
Mefatinib is an effective tyrosine kinase inhibitor with anti-tumor activity.
-
GC69459
Mefatinib free base
Mifanertinib
Mefatinib free base is an effective tyrosine kinase inhibitor with anti-tumor activity.
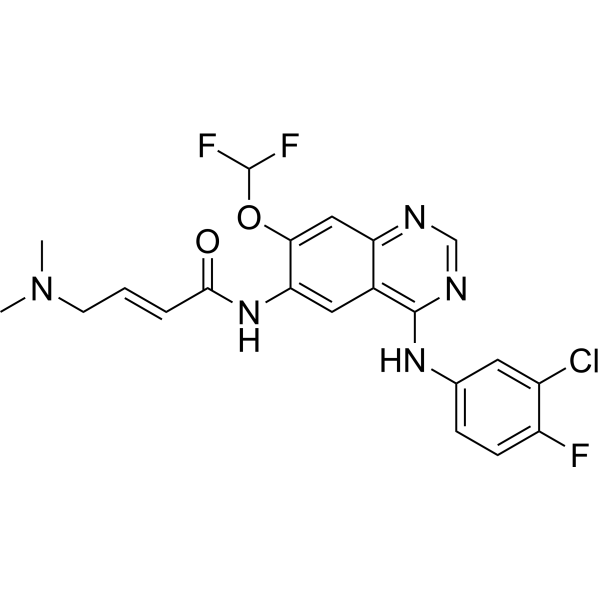
-
GC14951
Meleagrin
6-O-Methyloxaline
antibiotic
-
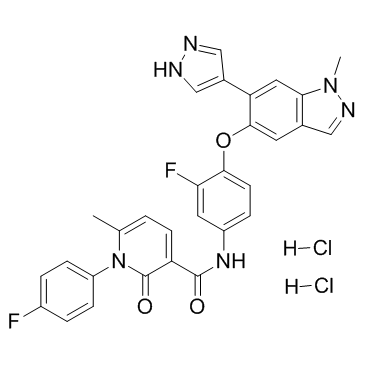
GC36585
Merestinib dihydrochloride
Merestinib dihydrochloride (LY2801653 dihydrochloride) is a potent, orally bioavailable c-Met inhibitor (Ki=2 nM) with anti-tumor activities. Merestinib dihydrochloride also has potent activity against MST1R (IC50=11 nM), FLT3 (IC50=7 nM), AXL (IC50=2 nM), MERTK (IC50=10 nM), TEK (IC50=63 nM), ROS1, DDR1/2 (IC50=0.1/7 nM) and MKNK1/2 (IC50=7 nM).
-
GC68018
MET kinase-IN-2

-
GC12069
Methyl 2,5-dihydroxycinnamate
Methyl 2,5dihydoxycinnamate
EGF receptor-associated tyrosine kinases inhibitor
-
GC36596
Methylnissolin
(–)-Methylnissolin
Methylnissolin (Astrapterocarpan), isolated from Astragalus membranaceus, inhibits platelet-derived growth factor (PDGF)-BB-induced cell proliferation with an IC50 of 10 μM.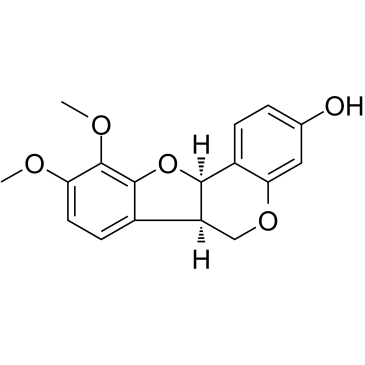
-
GC13598
MGCD-265
MGCD-265 is a potent and oral active inhibitor of c-Met and VEGFR2 tyrosine kinases, with IC50s of 29 nM and 10 nM, respectively. MGCD-265 has significant antitumor activity.

-
GC61516
MID-1
MID-1 is a disruptor of MG53-IRS-1 (Mitsugumin 53-insulin receptor substrate-1) interaction.

-
GC32819
Mirk-IN-1 (Dyrk1B/A-IN-1)
Dyrk1B/A-IN-1
Mirk-IN-1 (Dyrk1B/A-IN-1) is a potent inhibitor of Dyrk1B(Mirk kianse) and Dyrk1A with IC50 of 68±48 nM and 22±8 nM respectively.
-
GC16337
MK-2461
C-Met (WT/mutants) inhibitor

-
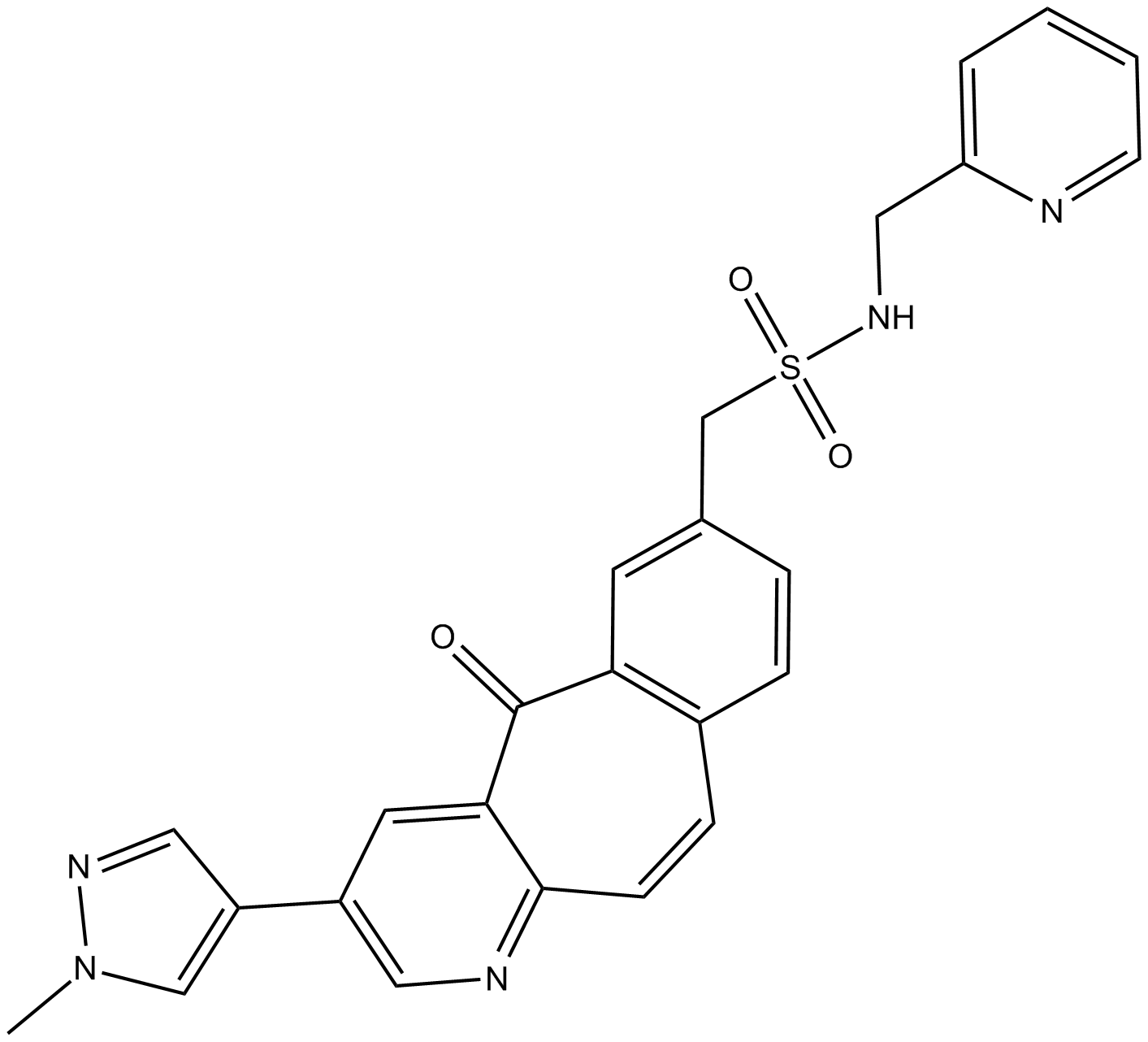
GC13140
MK-8033
MK 8033;MK8033
-
GC36625
MK-8033 hydrochloride
MK-8033 hydrochloride is an orally active ATP competitive c-Met/Ron dual inhibitor (IC50s: 1 nM (c-Met),7 nM (Ron)), with preferential binding to the activated kinase conformation. MK-8033 hydrochloride can be used in the research of cancers, such as breast and bladder cancers, non-small cell lung cancers (NSCLCs).

-
GC47687
ML-209
An RORγt antagonist
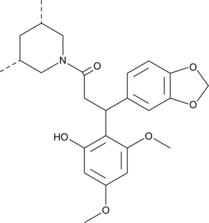
-
GC17582
ML347
LDN193719
BMP receptor inhibitor,potent and selective
-
GC30769
MLi-2
An LRRK2 inhibitor

-
GC10775
MLR 1023
CP 26,154, NSC 314335
MLR 1023 is a potent and selective allosteric activator of Lyn kinase with an EC50 of 63 nM.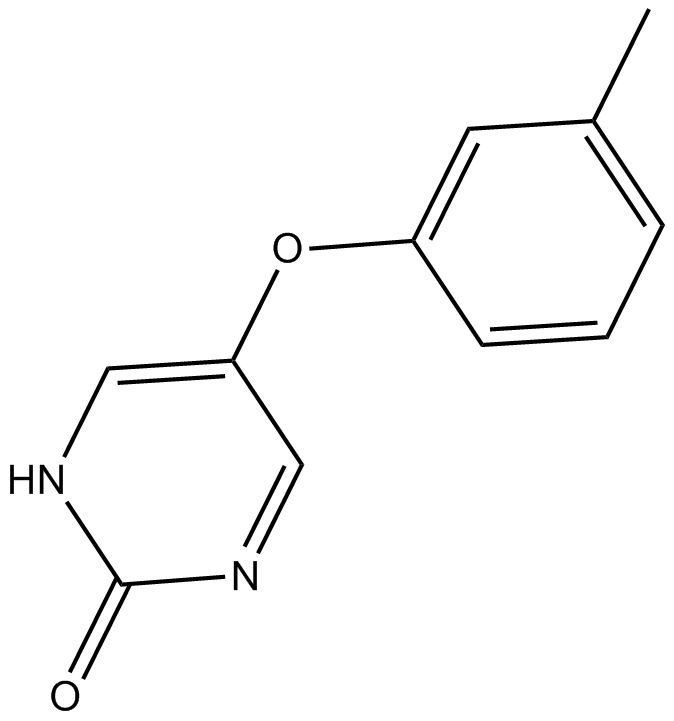
-
GC10048
MNS
3,4Methylenedioxyβnitrostyrene, NSC 10120, NSC 105303, NSC 170724, Syk Inhibitor III
Inhibitor of Src/Syk tyrosine kinases
-
GC47697
Mobocertinib
AP32788, TAK-788
An inhibitor of mutant EGFR and HER2 receptors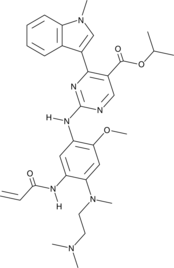
-
GC62160
Mobocertinib succinate
Mobocertinib (TAK-788) succinate is an orally active and irreversible EGFR/HER2 inhibitor. Mobocertinib succinate potently inhibits oncogenic variants containing activating EGFRex20ins mutations with selectivity over wild-type EGFR. Mobocertinib succinate can be used in NSCLC research.

-
GC13012
Motesanib

-
GC11336
Motesanib Diphosphate (AMG-706)
Motesanib
Motesanib Diphosphate (AMG-706) (AMG 706 Diphosphate) is a potent ATP-competitive inhibitor of VEGFR1/2/3 with IC50s of 2 nM/3 nM/6 nM, respectively, and has similar activity against Kit, and is approximately 10-fold more selective for VEGFR than PDGFR and Ret.
-
GC69496
MRL-871
MRL-871 (compound 3) is an effective inverse agonist of retinoic acid receptor-related orphan receptor gamma t (RORγt), with an IC50 of 264 nM. MRL-871 has a unique imidazole chemical structure and can effectively reduce the generation of IL-17a mRNA in EL4 cells.
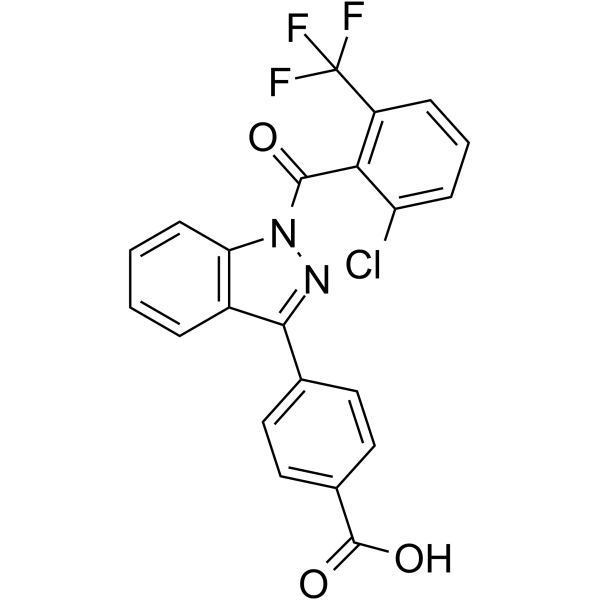
-
GC13525
MRS 4062 triethylammonium salt
P2Y4 receptor agonist
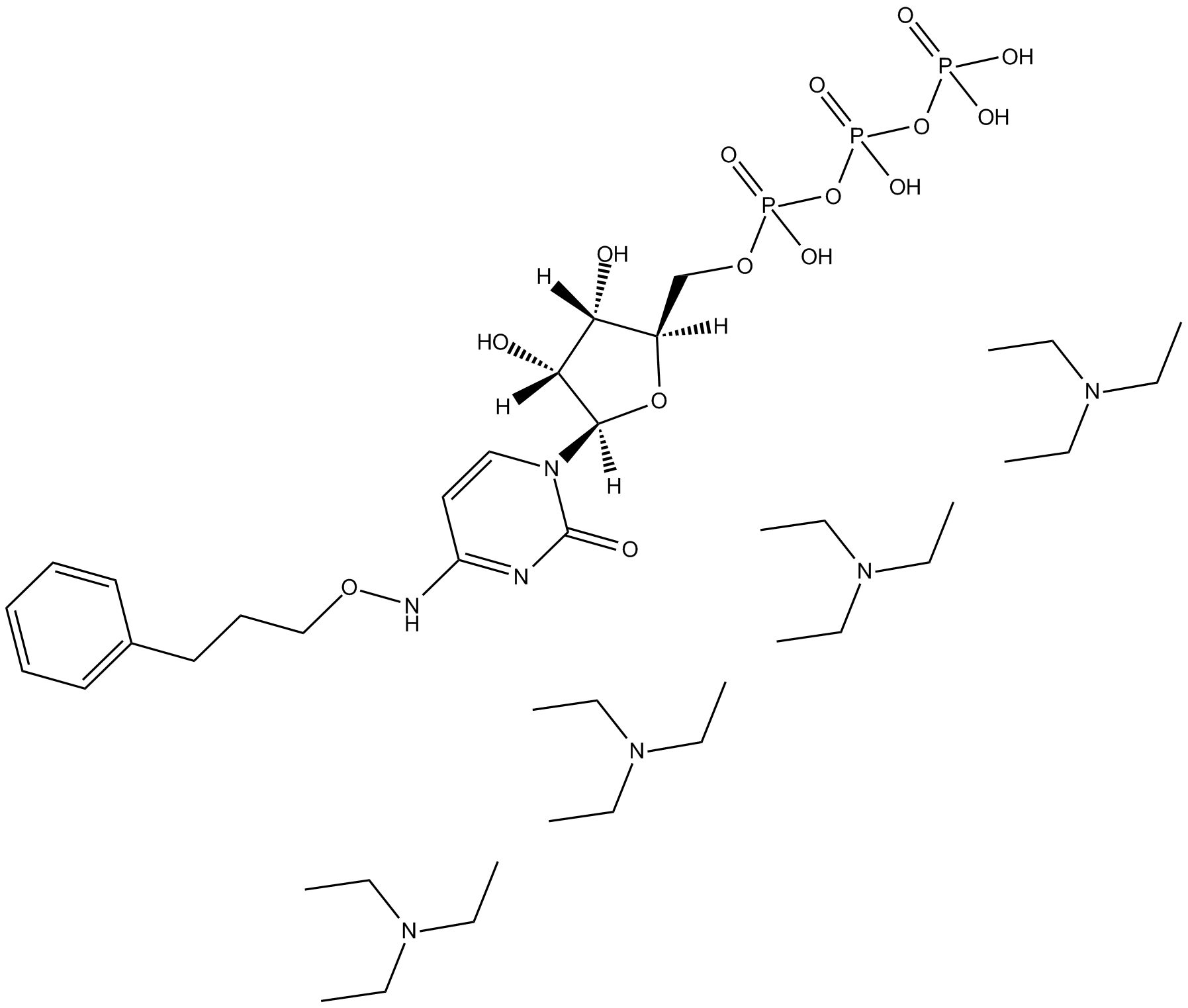
-
GC32769
MRX-2843 (UNC2371)
UNC2371
MRX-2843 (UNC2371) (UNC2371) is an orally active, ATP-competitive dual MERTK and FLT3 tyrosine kinases inhibitor (TKI) with enzymatic IC50s of 1.3 nM for MERTK and 0.64 nM for FLT3, respectively.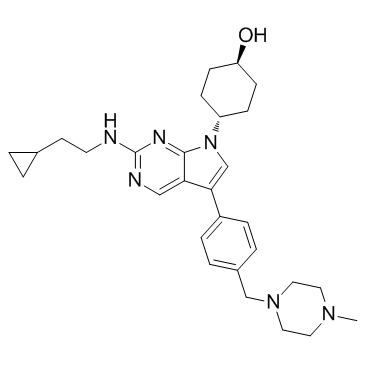
-
GC65243
MS4077
MS4077 is an anaplastic lymphoma kinase (ALK) PROTAC (degrader) based on Cereblon ligand, with a Kd of 37?nM for binding affinity to ALK.
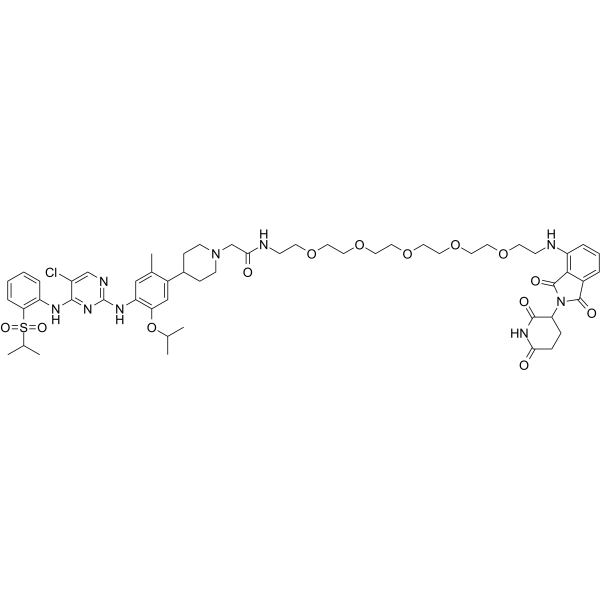
-
GC64966
MS4078
MS4078 is an anaplastic lymphoma kinase (ALK) PROTAC (degrader) based on Cereblon ligand, with a Kd of 19?nM for binding affinity to ALK.

-
GC15936
MSDC-0160
Mitoglitazone; CAY10415
mTOT-modulating insulin sensitizer
-
GC64431
MSDC-0602K
Azemiglitazone potassium
MSDC-0602K (Azemiglitazone potassium), a PPARγ-sparing thiazolidinedione (Ps-TZD), binds to PPARγ with the IC50 of 18.25 μM.
-
GC19256
MTX-211
Mol 211
MTX-211 is a dual inhibitor of EGFR and PI3K, used for the treatment of cancer and other diseases.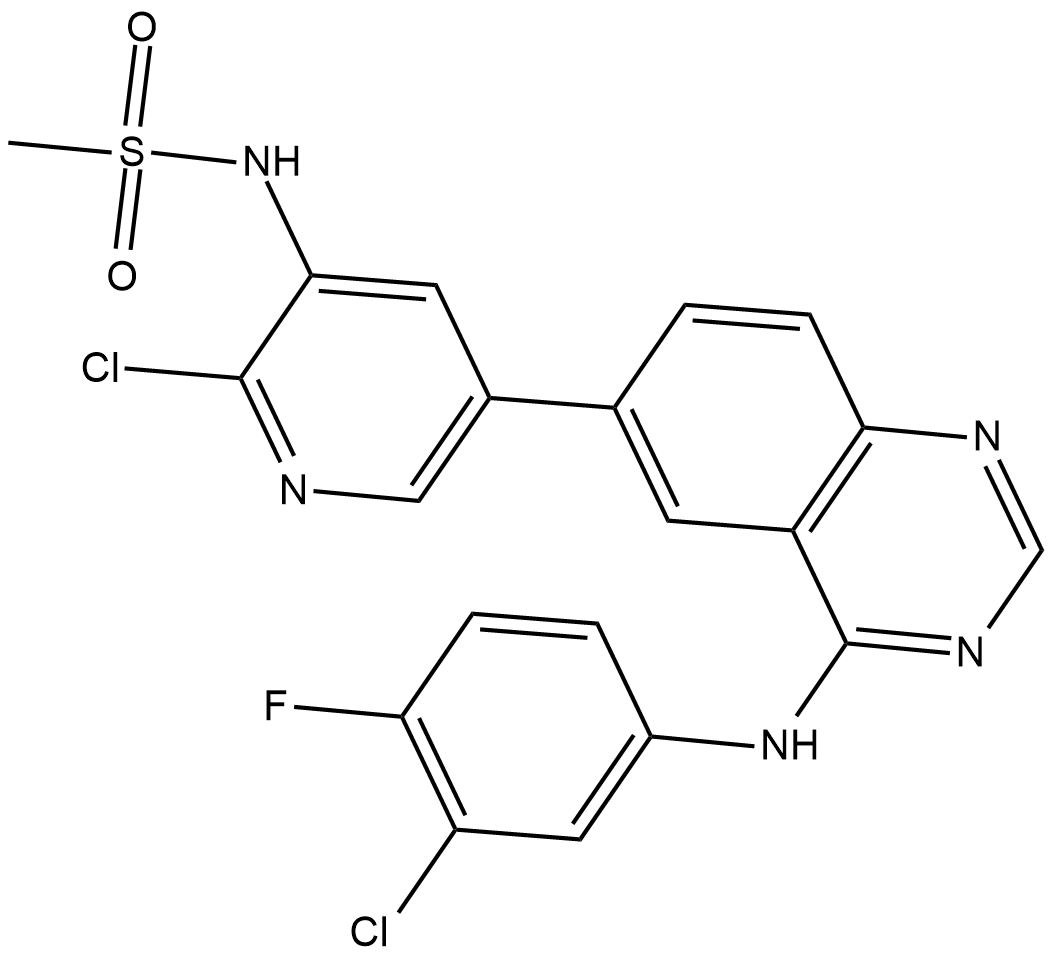
-
GC10250
Mubritinib (TAK 165)
TAK-165
Mubritinib (TAK 165) (TAK-165) is a potent and selective EGFR2/HER2 inhibitor with an IC50 of 6 nM.
-
GC11126
Mutant EGFR inhibitor
Selective Mutated EGFR inhibitor
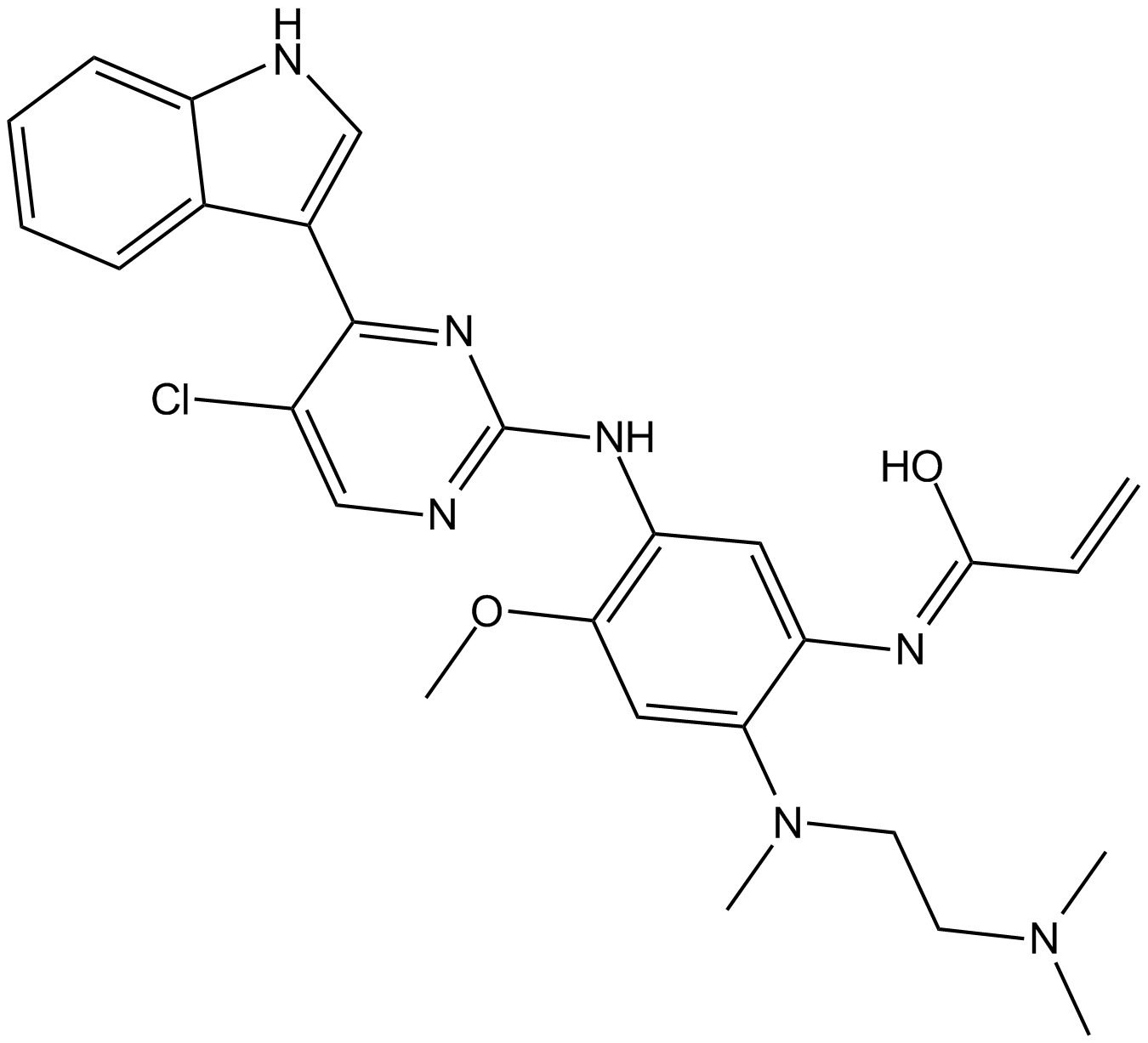
-
GC36666
Mutated EGFR-IN-1
Mutated EGFR-IN-1 (Osimertinib analog) is a useful intermediate for the inhibitors design for mutated EGFR, such as L858R EGFR, Exonl9 deletion activating mutant and T790M resistance mutant.
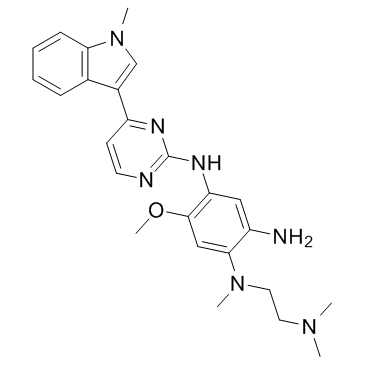
-
GC36667
Mutated EGFR-IN-2
Mutated EGFR-IN-2 (compound 91) is a mutant-selective EGFR inhibitor extracted from patent WO2017036263A1, which potently inhibits single-mutant EGFR (T790M) and double-mutant EGFR (including L858R/T790M (IC50=<1nM) and ex19del/T790M), and can suppress activity of single gain-of-function mutant EGFR (including L858R and ex19del) as well. Mutated EGFR-IN-2 shows anti-tumor antivity.

-
GC44263
Myrtillin
Delphinidin-3-β-D-glucoside chloride, Delphinidin-3-O-glucoside, Delphinin
Myrtillin (Delphinidin 3-o-glucoside) is a kind of anthocyanin monomer, which is mainly distributed in various plants and can be analyzed qualitatively and quantitatively by high performance liquid chromatography (HPLC) Mass spectrometry (Mass) and nuclear magnetic resonance (NMR)Myrtillin at 50 uM does not affect cell viability, also showing that delphinidin in association with lipopolysaccharide was able to induce MSC proliferation.

-
GC40567
N-(p-Coumaroyl) Serotonin
NSC 369503
N-(p-Coumaroyl) serotonin is an antioxidative phenolic naturally found in plants, including safflower seed and millet grain.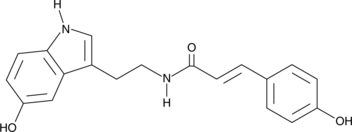
-
GC70279
N-Acetyl-5-hydroxytryptamine-d3
N-Acetyl-5-hydroxytryptamine-d3 is the deuterium labeled N-Acetyl-5-hydroxytryptamine.
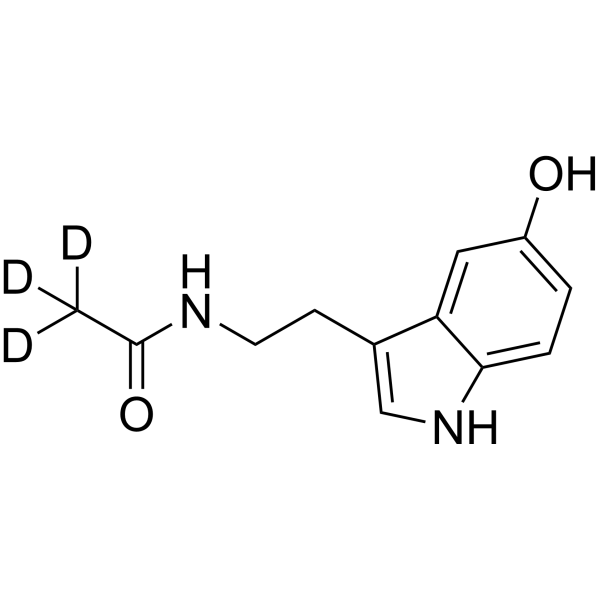
-
GC11526
N-Acetyl-O-phosphono-Tyr-Glu Dipentylamide
Phosphopeptide for binding to the src SH2 domain

-
GC12746
N-Acetyl-O-phosphono-Tyr-Glu-Glu-Ile-Glu
Phosphopeptide ligand for the src SH2 domain

-
GC15365
N-Acetylserotonin
N-Acetyl-5-hydroxytryptamine,NAS
N-Acetylserotonin is a Melatonin precursor, and that it can potently activate TrkB receptor.
-
GC49686
N-desmethyl Regorafenib N-oxide
An active metabolite of regorafenib

-
GC44290
NAADP (sodium salt)
Nicotinic acid adenine dinucleotide phosphate
Nicotinic acid adenine dinucleotide phosphate (NAADP) is a secondary messenger that induces calcium mobilization.

-
GC33045
NAMI-A
NAMI-A is a ruthenium-based drug characterised by the selective activity against tumour metastases, inhibits the adhesion and migration.

-
GC33022
Naquotinib (ASP8273)
ASP8273
Naquotinib (ASP8273) (ASP8273) is an orally available, mutant-selective and irreversible EGFR inhibitor; with IC50s of 8-33 nM toward EGFR mutants and 230 nM for EGFR.
-
GC32836
Naquotinib mesylate (ASP8273)
ASP8273 (mesylate)
Naquotinib mesylate (ASP8273) (ASP8273 mesylate) is an orally available, mutant-selective and irreversible EGFR inhibitor; with IC50s of 8-33 nM toward EGFR mutants and 230 nM for EGFR.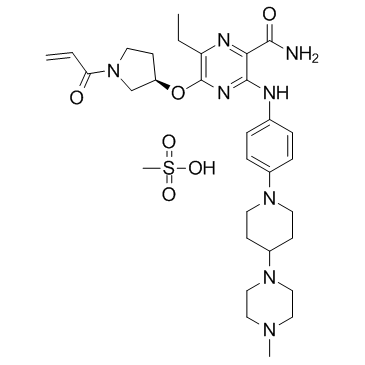
-
GC36699
Nazartinib mesylate
EGF816 mesylate
Nazartinib mesylate (EGF816 mesylate) is a novel, covalent mutant-selective EGFR inhibitor, with Ki and Kinact of 31 nM and 0.222 min?1 on EGFR(L858R/790M) mutant, respectively.
-
GC40623
NBI 31772
NBI 31772 is a nonpeptide ligand that releases insulin-like growth factor-1 (IGF-1) from its binding protein (IGFBP; Kis = 1-24 nM for the six human subtypes of IGFBP).

-
GC61111
NBI-31772 hydrate
NBI-31772 hydrate is a potent inhibitor of interaction between insulin-like growth factor (IGF) and IGF-binding proteins (IGFBPs).

-
GC10644
Neoruscogenin
25(27)-Dehydroruscogenin
nuclear receptor RORα agonist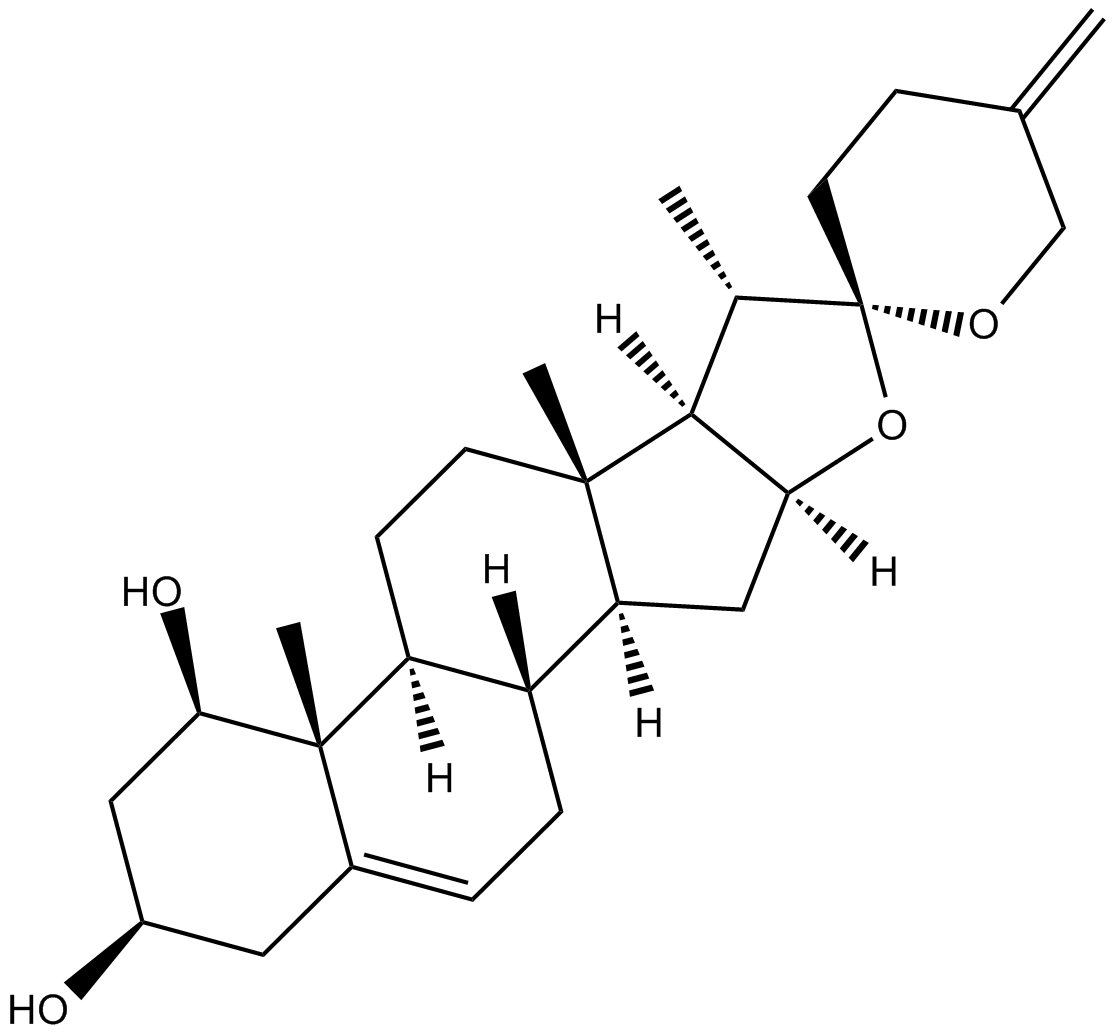
-
GC10362
Neratinib (HKI-272)
HKI-272;HKI272;HKI 272
Neratinib (HKI-272) (HKI-272) is an orally available, irreversible, highly selective HER2 and EGFR inhibitor with IC50s of 59 nM and 92 nM, respectively.
-
GC47771
NG 25 (hydrochloride hydrate)
An inhibitor of MAP4K2 and TAK1
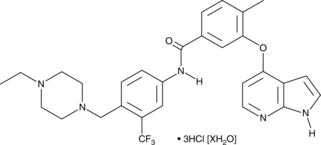
-
GC60270
Nilotinib D6
An internal standard for the quantification of nilotinib
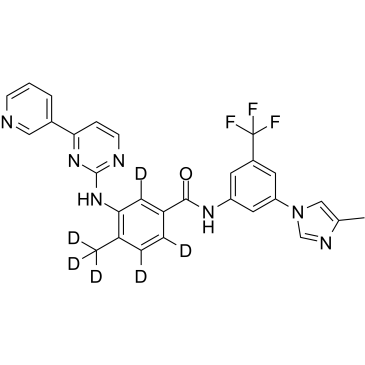
-
GC25669
Nilotinib hydrochloride
AMN-107 HCl
Nilotinib hydrochloride (AMN-107) is the hydrochloride salt form of nilotinib, an orally bioavailable Bcr-Abl tyrosine kinase inhibitor with antineoplastic activity.
-
GC14237
Nilotinib monohydrochloride monohydrate
A Bcr-Abl inhibitor

-
GC14129
Nilotinib(AMN-107)
AMN107
A Bcr-Abl inhibitor
-
GC62601
Nimotuzumab
Nimotuzumab is a humanized IgG1 monoclonal antibody targeting EGFR with a KD of 0.21 nM. Nimotuzumab is directed against the extracellular domain of the EGFR blocking the binding to its ligands. Nimotuzumab, a strong antitumor drug, is cytolytic on target tumors by its capacity to cause antibody dependent cell mediated cytotoxicity (ADCC) and complement dependent cytotoxicity (CDC).

-
GC18211
Ningetinib
A multi-kinase inhibitor

-
GC36744
Ningetinib Tosylate
Ningetinib Tosylate is a potent, orally bioavailable small molecule tyrosine kinase inhibitor (TKI) with IC50s of 6.7, 1.9 and <1.0 nM for c-Met, VEGFR2 and Axl, respectively.

-
GC11705
Nintedanib (BIBF 1120)
Vargatef
A VEGFR, FGFR, and PDGFR inhibitor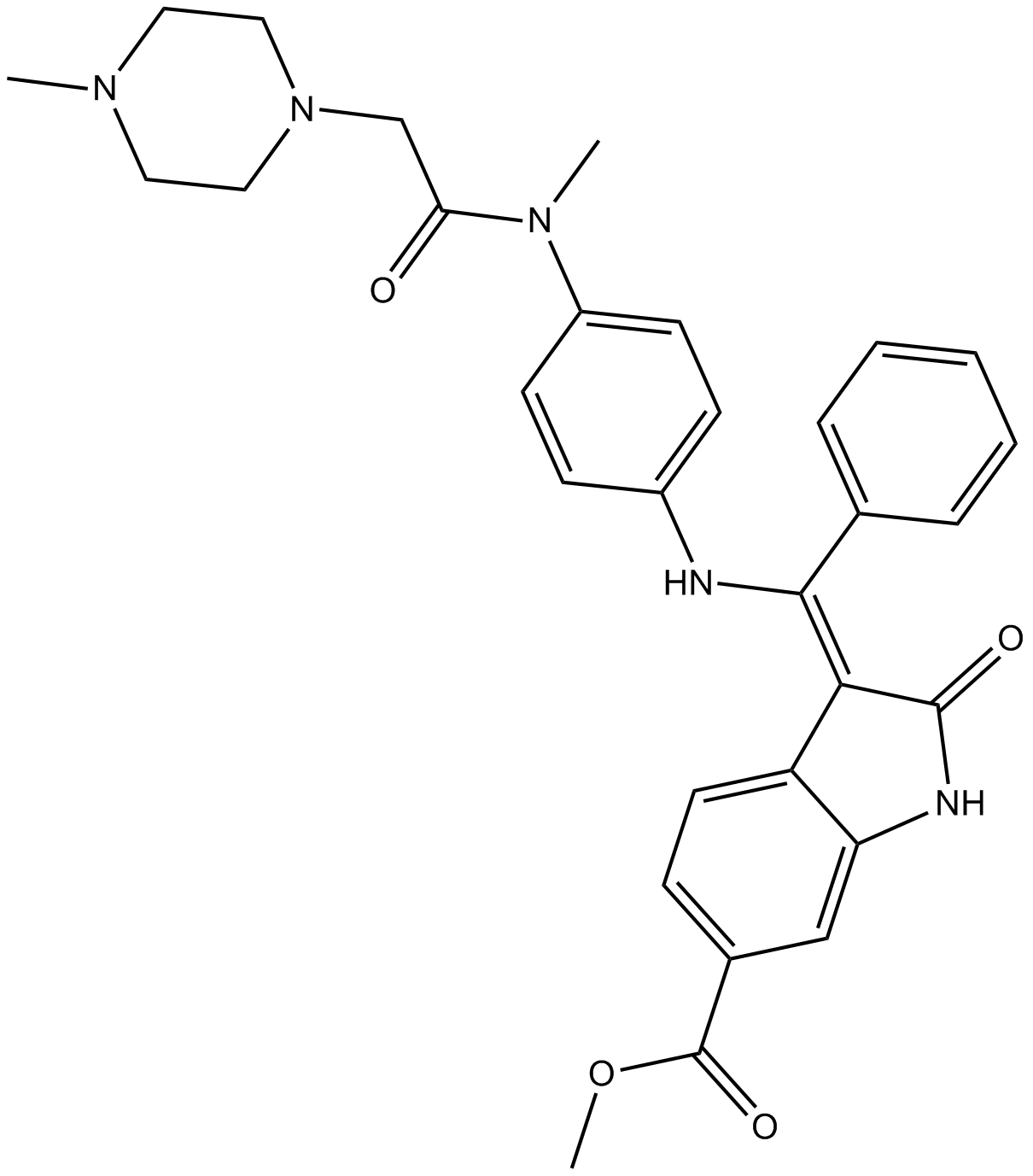
-
GC36745
Nintedanib esylate
Nintedanib
Nintedanib esylate, as a kinase inhibitor, used for the treatment of non-small cell lung cancer suffered from first-pass metabolism which resulted in low oral bioavailability (~ 4.7%).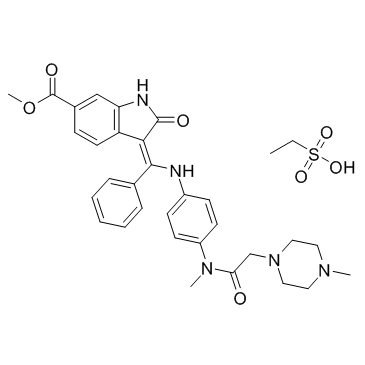
-
GN10325
Nobiletin
NSC 76751, NSC 618903

-
GC14075
Nocodazole
NSC 238159, Oncodazole, R 17934
Nocodazole is an anti-mitotic drug and a rapid and reversible microtubule polymerization inhibitor. It inhibits Abl, Abl (E255K), and Abl (T315I) in cell-free assays with IC50 values of 0.21μM, 0.53μM, and 0.64μM, respectively.
-
GC50152
Norleual
Highly potent HGF/c-MET inhibitor; also AT4 antagonist

-
GC69584
Norleual TFA
Norleual TFA is a type IV angiotensin (Ang) similar substance and a hepatocyte growth factor (HGF)/c-Met inhibitor with an IC50 of 3 pM. It is also an AT4 antagonist with strong anti-angiogenic activity.

-
GC14488
NPS-1034
MET inhibitor

-
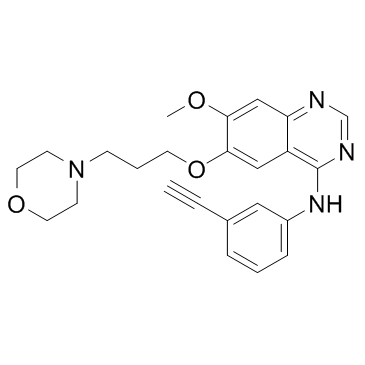
GC33131
NRC-2694
NRC-2694 is an epidermal growth factor receptor (EGFR) antagonist with anti-cancer and anti-proliferative properties.
-
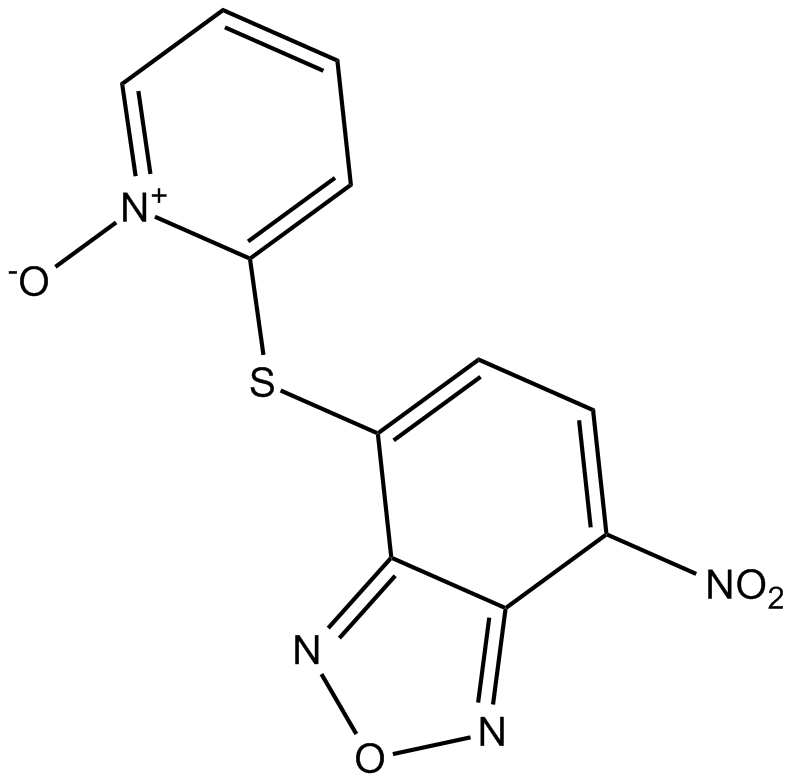
GC14103
NSC228155
EGFR activator
-
GC69596
NSC689857
NSC689857 is an effective inhibitor of EGFR and SCFSKP2, with an IC50 of 36 μM against Skp2-Cks1. NSC689857 can inhibit phosphorylation of p27 (IC50=30 μM). NSC689857 exhibits varying activity in different types of cancer, with higher resistance activity against leukemia cell lines compared to other cancer cells.

-
GC12712
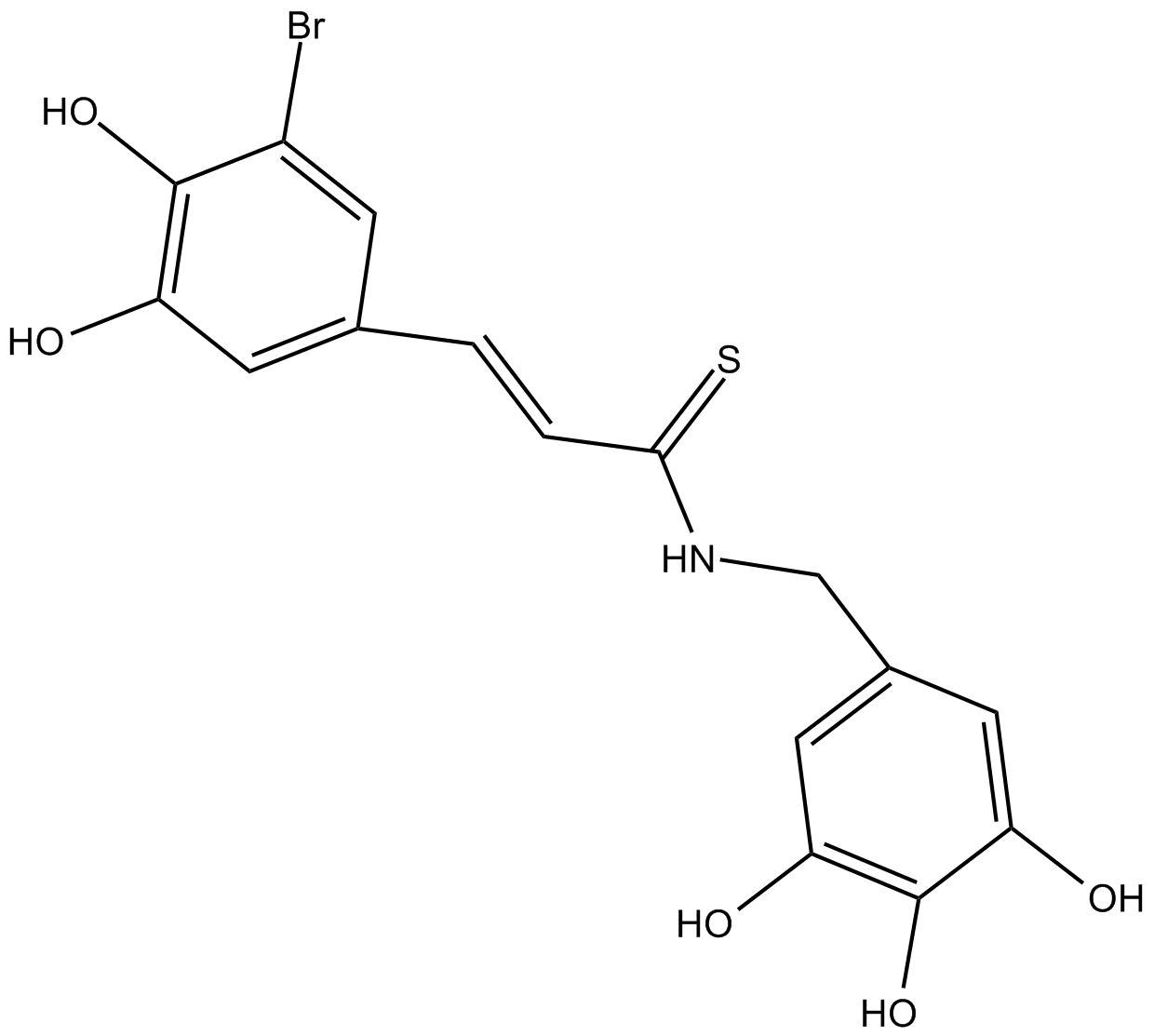
NT157
IRS-1/2 inhibitor, inhibits IGF-1R and STAT3 signaling pathway
-
GC69599
NT219
NT219 is an effective dual inhibitor of insulin receptor substrate 1/2 (IRS1/2) and STAT3. IRS1/2 and STAT3 are major signaling pathways regulated by various oncogenes. NT219 affects the degradation of IRS1/2 and inhibits the phosphorylation of STAT3. NT219 has potential for researching cancer diseases.

-
GC10720
NTR 368
cytoplasmic peptide of the neurotrophin receptor p75NTR

-
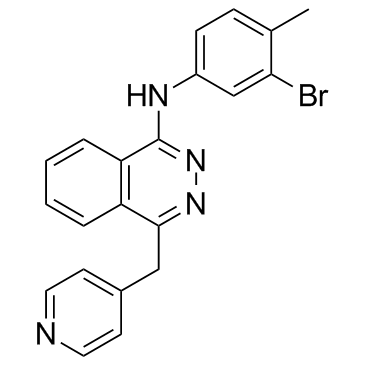
GC34126
NVP-ACC789 (ACC-789)
NVP-ACC789 (ACC-789) is an inhibitor of human VEGFR-1, VEGFR-2 (mouse VEGFR-2), VEGFR-3 and PDGFR-β with IC50s of 0.38, 0.02 (0.23), 0.18, 1.4 μM, respectively.
-
GC14310
NVP-ADW742
ADW742, GSK552602A
Selective IGF-1R inhibitor
-
GC12963
NVP-AEW541
AEW541
IGF-IR inhibitor, novel, potent and selective
-
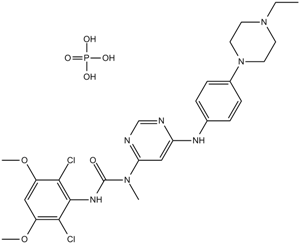
GC16028
NVP-BGJ398 phosphate
-
GC14332
NVP-BHG712
EphB4 inhibitor,potent and selective
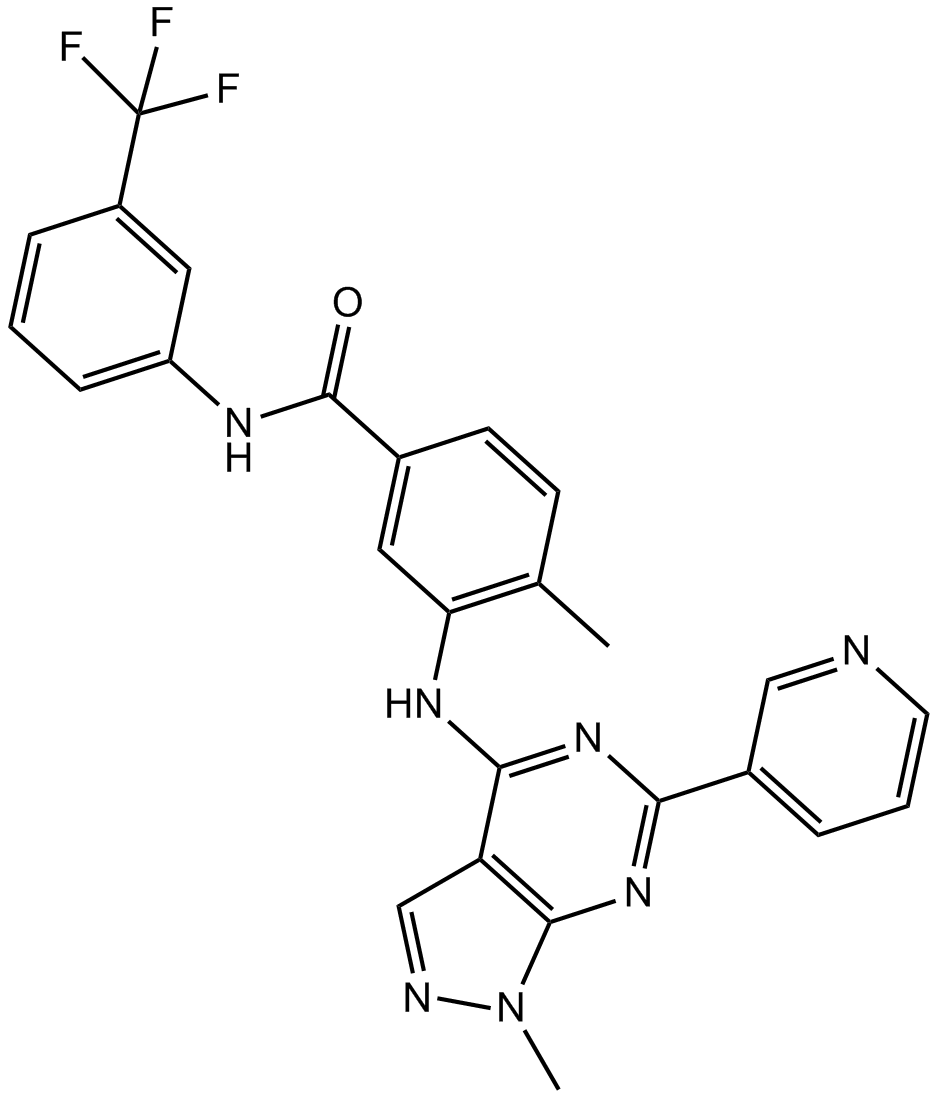
-
GC36782
NVP-BHG712 isomer
NVP-BHG712 isomer, a regioisomer of NVP-BHG712, shows conserved non-bonded binding to EPHA2 and EPHB4.
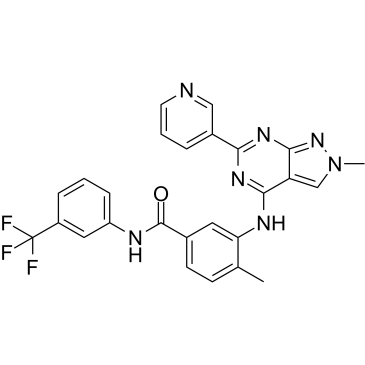
-
GC16972
NVP-BVU972
C-Met inhibitor,potent and selective

-
GC44491
O-Desmethyl Gefitinib
O-Desmethyl gefitinib is the major metabolite of gefitinib in human plasma, formed by the cytochrome P450 isoform CYP2D6.

-
GC68321
O-Desmethyl gefitinib-d8

-
GC64950
ODM-203
ODM-203 is an orally active and selective FGFR/VEGFR inhibitor with IC50 values of 6, 11, 16, 5, 9, 26 and 35 nM for FGFR3/1/2 and VEGFR3/2/1/4, respectively. ODM-203 has strong anti-tumour activity and activates immune responses in the tumour microenvironment.

-
GC32502
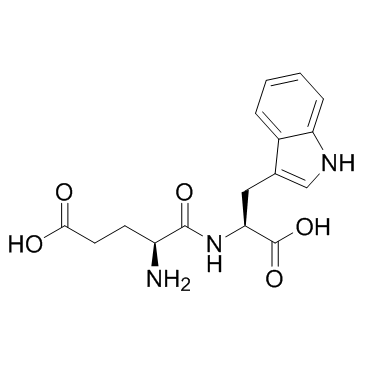
Oglufanide (H-Glu-Trp-OH)
Oglufanide (H-Glu-Trp-OH) (H-Glu-Trp-OH) is a dipeptide immunomodulator isolated from calf thymus.
-
GC69619
Olaratumab
IMC-3G3; LY3012207
Olaratumab (IMC-3G3; LY3012207) is a human monoclonal IgG1 antibody that targets platelet-derived growth factor receptor alpha (PDGFRα) and has anti-tumor activity.

-
GC15370
Olmutinib (HM61713, BI 1482694)
BI-1482694, Olmutinib
Olmutinib (HM61713, BI 1482694) (HM61713; BI-1482694) is an orally active and irreversible third EGFR tyrosine kinase inhibitor that binds to a cysteine residue near the kinase domain. Olmutinib (HM61713, BI 1482694) is used for NSCLC.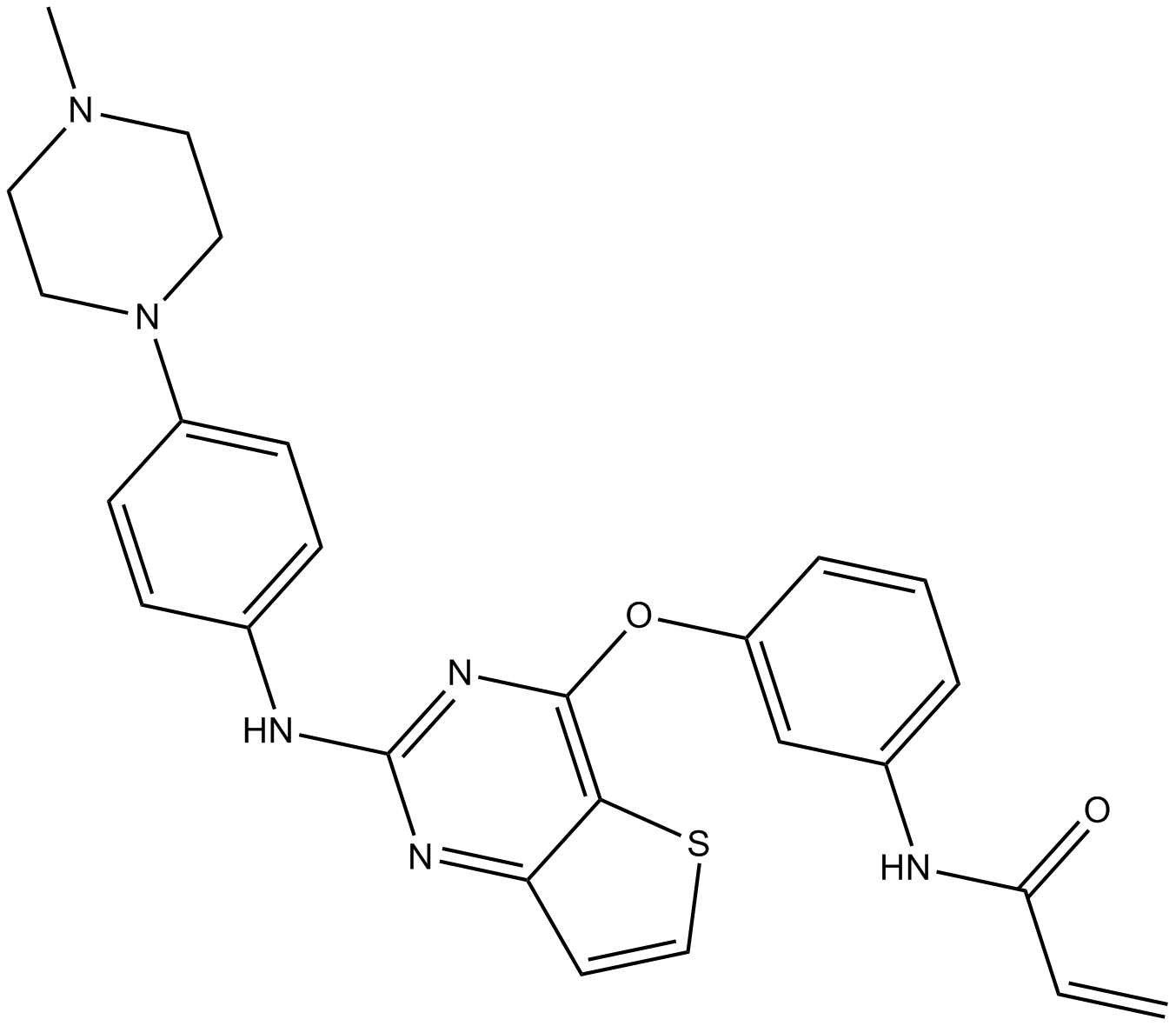
-
GC62207
Olverembatinib
GZD824; HQP1351
Olverembatinib (GZD824) is a potent and orally active pan-Bcr-Abl inhibitor. Olverembatinib potently inhibits a broad spectrum of Bcr-Abl mutants. Olverembatinib strongly inhibits native Bcr-Abl and Bcr-AblT315I with IC50s of 0.34 nM and 0.68 nM, respectively. Olverembatinib has antitumor activity.
-
GC36807
ON 146040
ON 146040 is a potent PI3Kα and PI3Kδ (IC50≈14 and 20 nM, respectively) inhibitor. ON 146040 also inhibits Abl1 (IC50<150 nM).



