Tyrosine Kinase
- IGFBR(1)
- Bcr-Abl(65)
- Ack1(2)
- Axl(6)
- ALK(54)
- BMX Kinase(3)
- Broad Spectrum Protein Kinase Inhibitor(10)
- c-FMS(33)
- c-Kit(56)
- c-MET(81)
- c-RET(0)
- CSF-1R(3)
- DDR1/DDR2 Receptor(1)
- EGFR(229)
- EphB4(1)
- FAK(33)
- FGFR(87)
- FLT3(85)
- HER2(12)
- IGF1R(30)
- Insulin Receptor(41)
- IRAK(25)
- ITK(10)
- Lck(1)
- LRRK2(22)
- PDGFR(93)
- PTKs/RTKs(3)
- Pyk2(6)
- ROR(38)
- Spleen Tyrosine Kinase (Syk)(31)
- Src(90)
- Tie-2 (3)
- Trk(35)
- VEGFR(169)
- Kinase(0)
- Discoidin Domain Receptor(14)
- DYRK(26)
- Ephrin Receptor(12)
- ROS(14)
- RET(28)
- TAM Receptor(28)
Products for Tyrosine Kinase
- Cat.No. Product Name Information
-
GC17473
Pelitinib (EKB-569)
EKB569
Pelitinib (EKB-569) (EKB-569;WAY-EKB 569) is an irreversible inhibitor of EGFR with an IC50 of 38.5 nM; also slightly inhibits Src, MEK/ERK and ErbB2 with IC50s of 282, 800, and 1255 nM, respectively.
-
GC32915
Pemigatinib
INCB054828
An FGFR inhibitor
-
GC60283
Pentagamavunon-1
PGV-1
Pentagamavunon-1 (PGV-1), a Curcumin analog with oral activity, targets on several molecular mechanisms to induce apoptosis including inhibition of angiogenic factors cyclooxygenase-2 (COX-2) and vascular endothelial growth factor (VEGF). PGV-1 inhibits NF-κB activation.
-
GC47938
Pericosine A
(+)-Pericosine A
A fungal metabolite with anticancer activity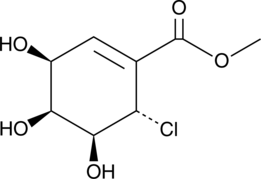
-
GC34210
Pertuzumab (Anti-Human HER2, Humanized Antibody)
Pertuzumab (Anti-Human HER2, Humanized Antibody), the first of a new class of agents designated as HER dimerisation inhibitors, is a humanised IgG1 monoclonal antibody (mAb) that sterically binds domain II of the erbB2 receptor.

-
GC69690
Petosemtamab
MCLA 158
Petosemtamab (MCLA 158) is a monoclonal antibody (mAb) that targets both EGFR (Kd: 0.22 nM) and LGR5 (Kd: 0.86 nM). Petosemtamab blocks EGFR signaling and receptor degradation in LGR5+ cancer cells. It can be used for research on solid tumors such as head and neck squamous cell carcinoma (HNSCC), metastatic colorectal cancer (CRC), etc.

-
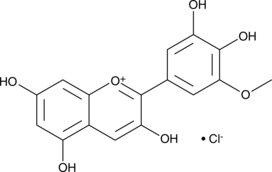
GC44605
Petunidin (chloride)
Petunidol
Petunidin is an O-methylated anthocyanidin derived from delphinidin that imparts blue-red pigments to flowers, fruits, and red wine.
-
GC12222
Pexidartinib (PLX3397)
PLX3397
Pexidartinib (PLX3397) (PLX-3397) is a potent, orally active, selective, and ATP-competitive colony stimulating factor 1 receptor (CSF1R or M-CSFR) and c-Kit inhibitor, with IC50s of 20 and 10 nM, respectively. Pexidartinib (PLX3397) (PLX-3397) exhibits 10- to 100-fold selectivity for c-Kit and CSF1R over other related kinases. Pexidartinib (PLX3397) (PLX-3397) induces cell apoptosis and has anti-tumor activity.
-
GC34708
Pexidartinib hydrochloride
Pexidartinib hydrochloride (PLX-3397 hydrochloride) is a potent, orally active, selective, and ATP-competitive colony stimulating factor 1 receptor (CSF1R or M-CSFR) and c-Kit inhibitor, with IC50s of 20 and 10 nM, respectively. Pexidartinib hydrochloride exhibits 10- to 100-fold selectivity for c-Kit and CSF1R over other related kinases. Pexidartinib hydrochloride induces cell apoptosis and has anti-cancer activity.

-
GC50346
PF 06273340
Potent and selective pan-Trk inhibitor; peripherally restricted

-
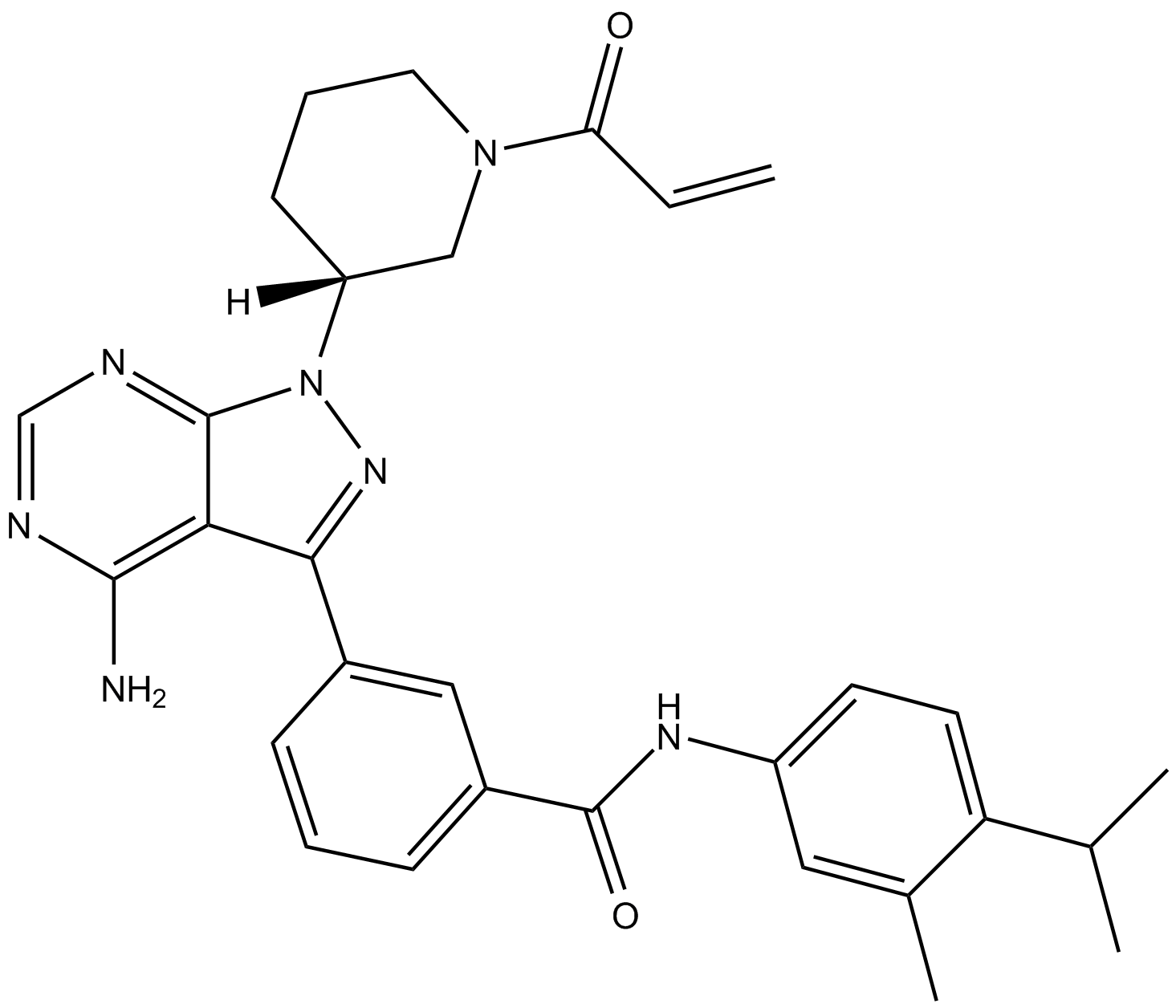
GC17630
PF 06465469
inhibitor of interleukin-2 inducible T cell kinase (ITK) and Bruton's tyrosine kinase (BTK)
-
GC14767
PF-00562271
PF-562271;PF00562271;PF62271
PF-562271 (VS-6062) besylate is a potent ATP-competitive, reversible inhibitor of FAK and Pyk2 kinase, with an IC50 of 1.5 nM and 13 nM, respectively.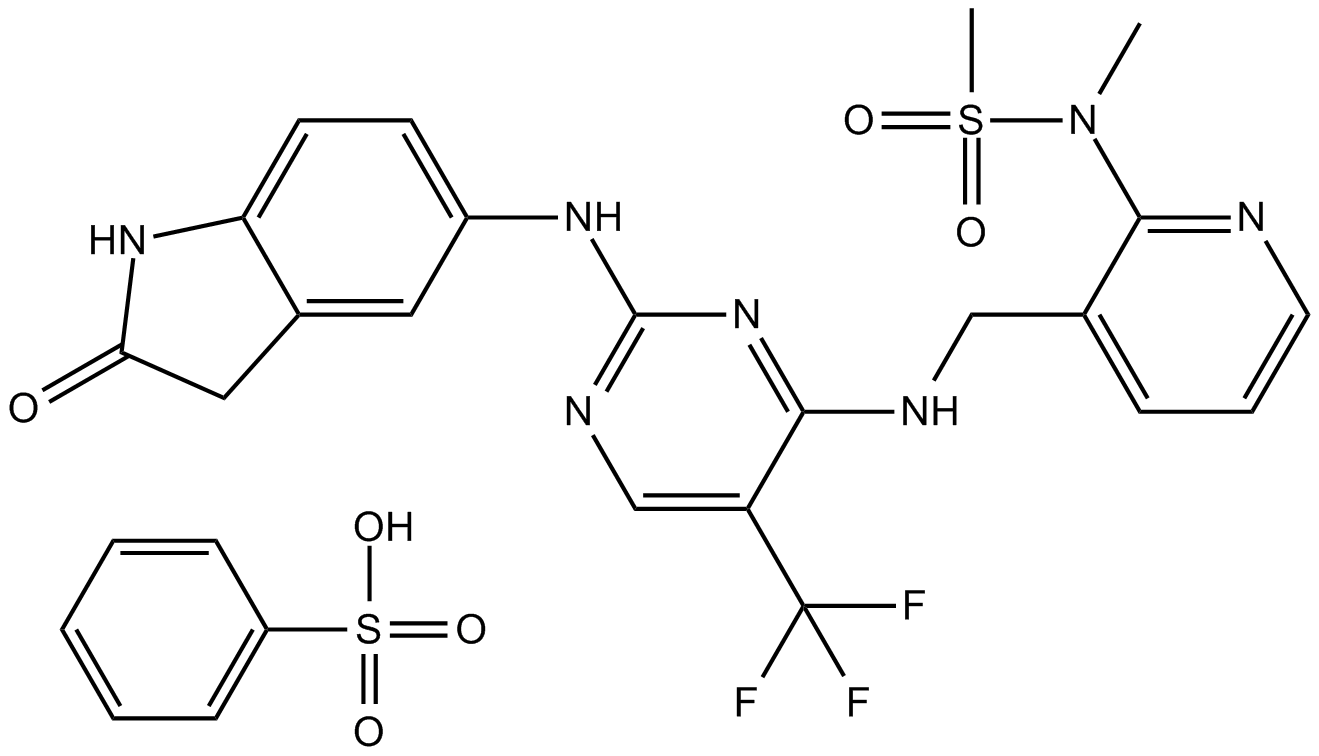
-
GC18074
PF-03814735
PF-03814735 is a potent, orally bioavailable, reversible inhibitor of both Aurora1 and Aurora2 kinases with IC50 values of 0.8nM and 5nM, respectively [1].

-
GC12729
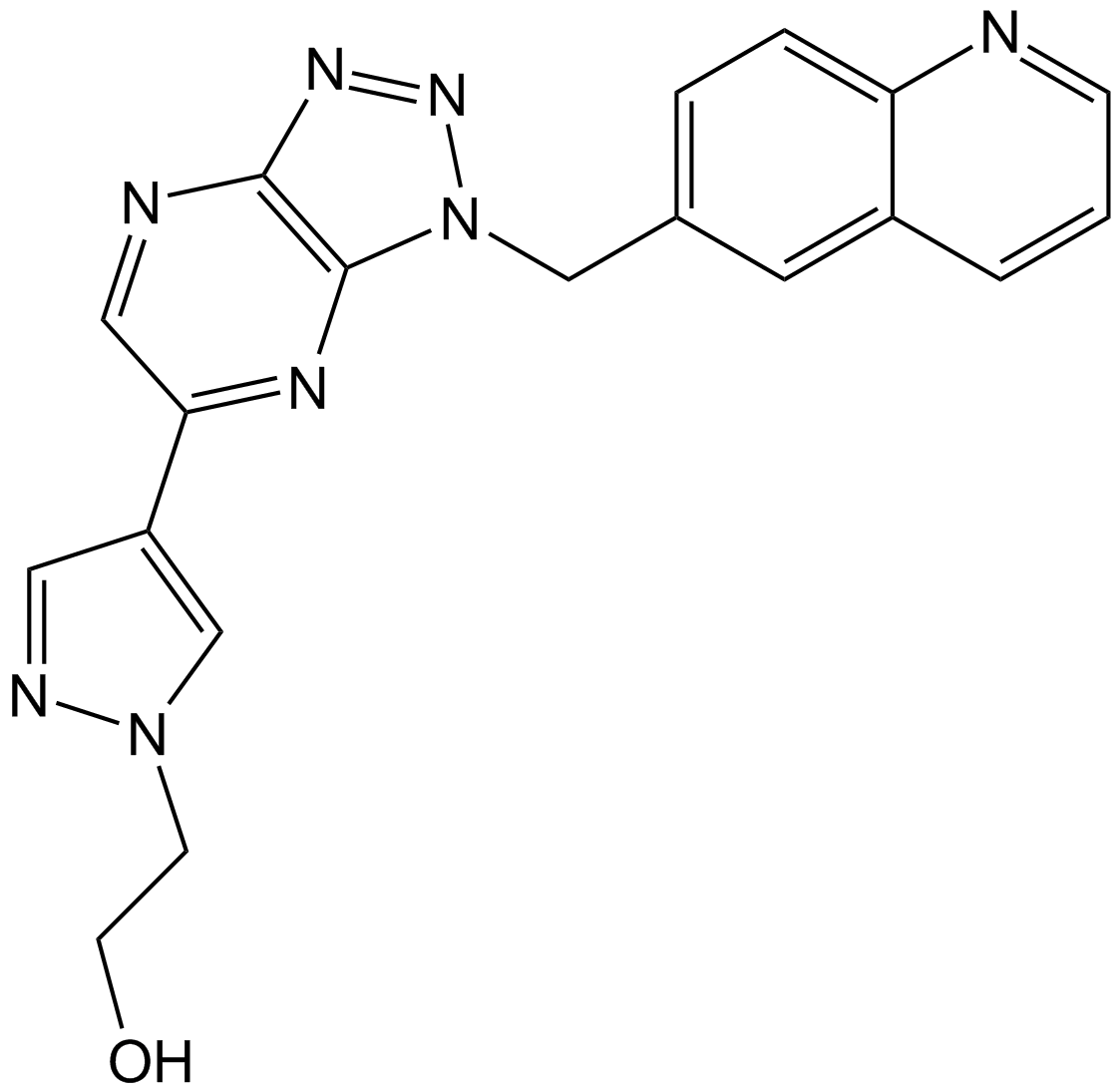
PF-04217903
C-Met inhibitor,selective and ATP-competitive
-
GC15733
PF-04217903 methanesulfonate
PF04217903 mesylate
A c-Met inhibitor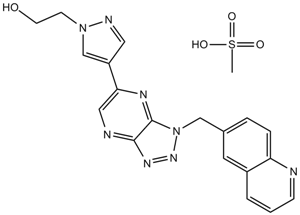
-
GC31495
PF-05231023
Mal-PEG2-AZD
PF-05231023 is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.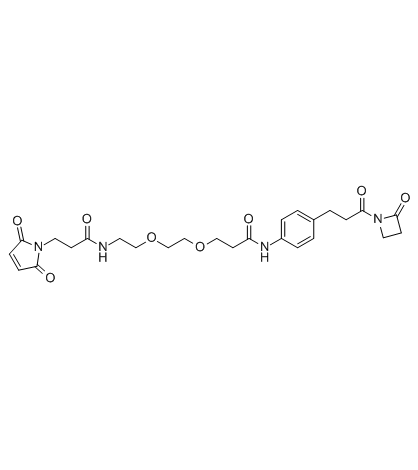
-
GC13850
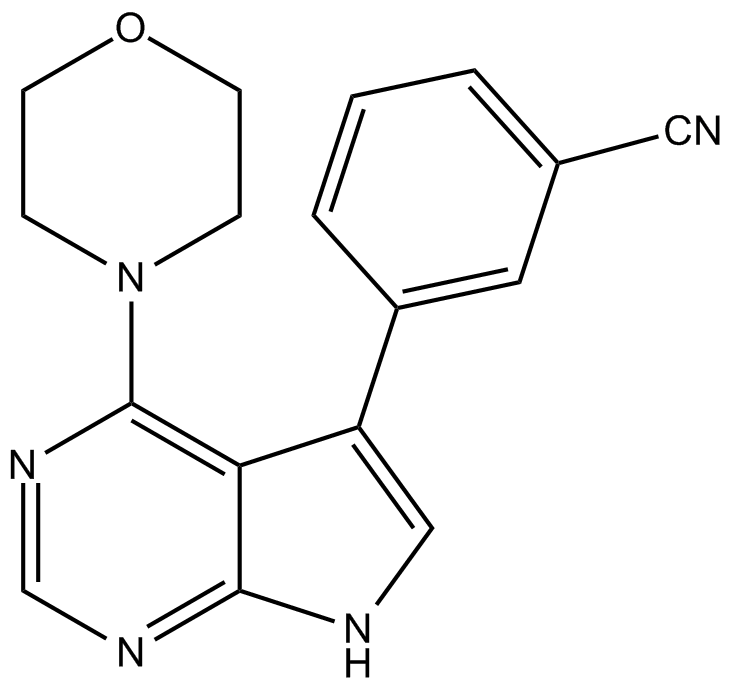
PF-06447475
LRRK2 inhibitor
-
GC64566
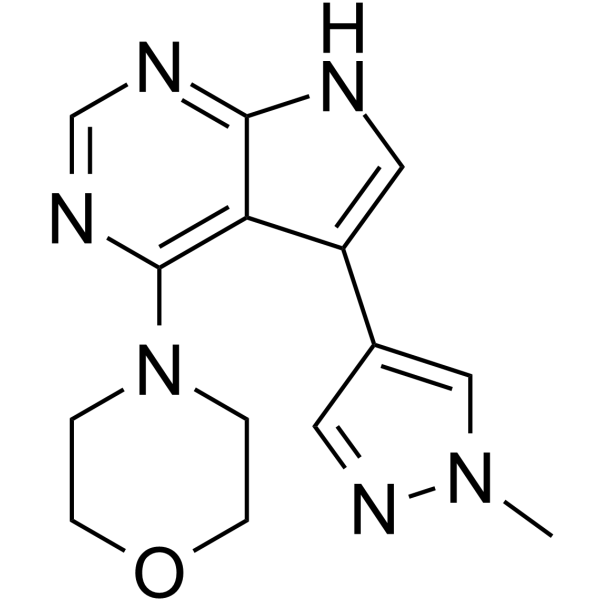
PF-06454589
PF-06447475 is a highly potent, selective, brain penetrant LRRK2 kinase inhibitor with IC50 values of 3 nM and 11 nM for WT LRRK and G2019S LRRK2, respectively.
-
GC64506
PF-06456384 trihydrochloride
PF-06447475 trihydrochloride is a highly potent, selective, brain penetrant LRRK2 kinase inhibitor with IC50 values of 3 nM and 11 nM for WT LRRK and G2019S LRRK2, respectively.

-
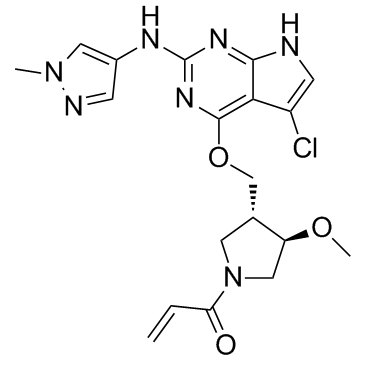
GC32927
PF-06459988
PF-06459988 is an orally activity, irreversible and mutant-selective inhibitor of EGFR mutant forms. PF-06459988 demonstrates high potency and specificity to the T790M-containing double mutant EGFRs. PF-06459988 can be used for the research of cancer.
-
GC14794
PF-06463922
Lorlatinib
PF-06463922 (PF-06463922) is a selective, orally active, brain-penetrant and ATP-competitive ROS1/ALK inhibitor. PF-06463922 has Kis of <0.025 nM, <0.07 nM, and 0.7 nM for ROS1, wild type ALK, and ALKL1196M, respectively. PF-06463922 has anticancer activity.
-
GC34710
PF-06747711
PF-06747711 is a potent, selective, and orally active retinoic acid receptor-related orphan C2 (RORC2, also known as RORγt) inverse agonist, with an IC50 of 4.1 nM.

-
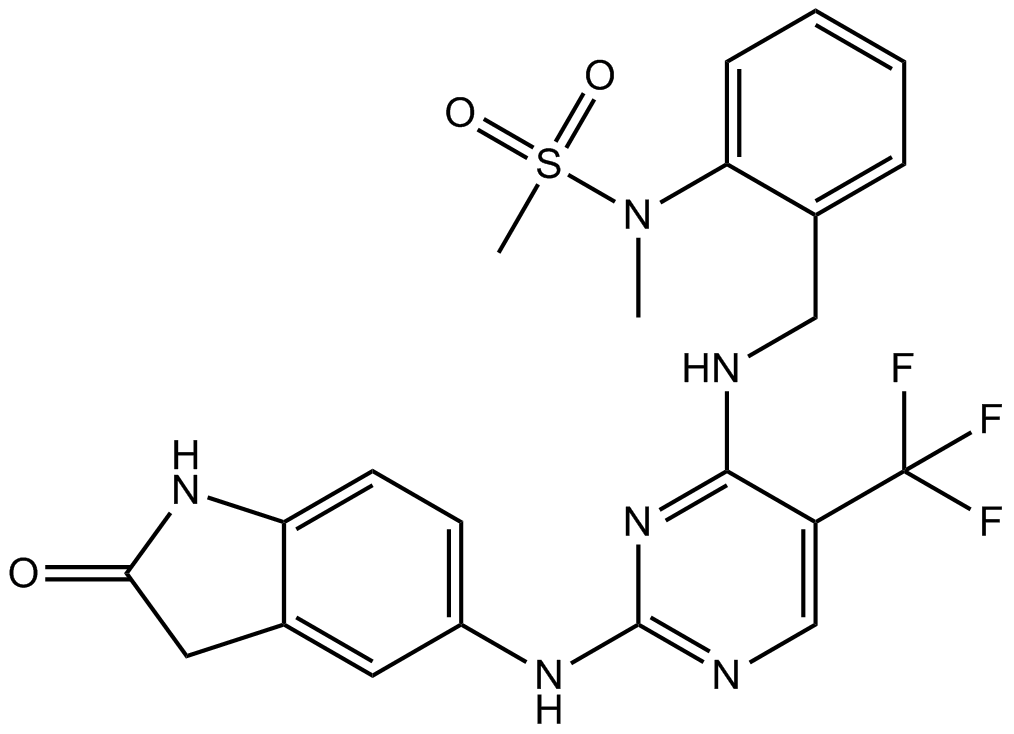
GC14407
PF-431396
Pyk2 and FAK inhibitor
-
GC61852
PF-4618433
PF-4618433 is a potent and selective PYK2 inhibitor, with an IC50 of 637 nM.

-
GC15380
PF-562271
PF562271;PF 562271
ATP-competitive FAK inhibitor, reversible
-
GC10810
PF-562271 HCl
PF562271 HCl;PF 562271 HCl
PF-562271 (VS-6062) hydrochloride is a potent, ATP-competitive and reversible FAK and Pyk2 kinase inhibitor with IC50s of 1.5 nM and 13 nM, respectively.
-
GC11107
PF-573228
FAK Inhibitor II, Focal Adhesion Kinase Inhibitor II
ATP-competitive FAK inhibitor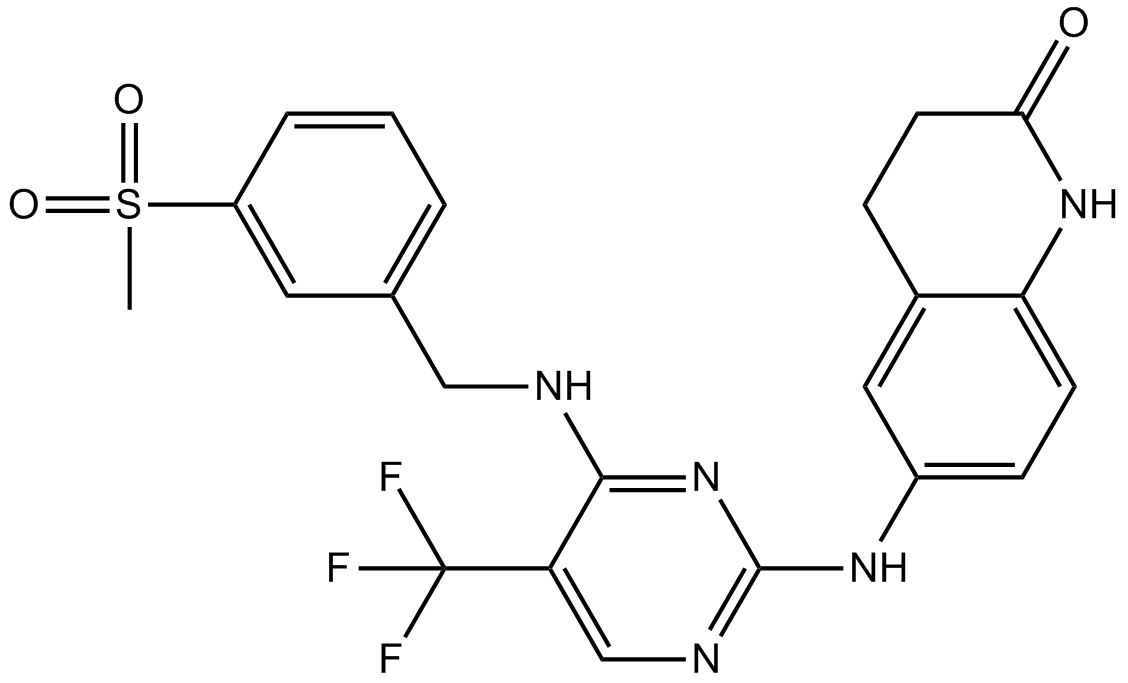
-
GC44613
PF-6274484
PF-6274484 is an inhibitor of the EGF receptor (EGFR; IC50s = 0.18 and 0.14 nM for wild-type EGFR and inhibitor-resistant EGFRL858R/T790M, respectively).

-
GC65009
PF-6683324
Trk-IN-4
PF-6683324 (Trk-IN-4) is a potent pan-Trk inhibitor in cell-based assays withIC50s of 1.9 nM, 2.6 nM and 1.1 nM for TrkA, TrkB and TrkC, respectively.
-
GC19287
PF06650833
PF-06650833
PF06650833 (PF-06650833) is a potent, selective and orally active inhibitor of interleukin-1 receptor associated kinase 4 (IRAK4) with IC50s of 0.2 and 2.4 nM in the cell and PBMC assay, respectively.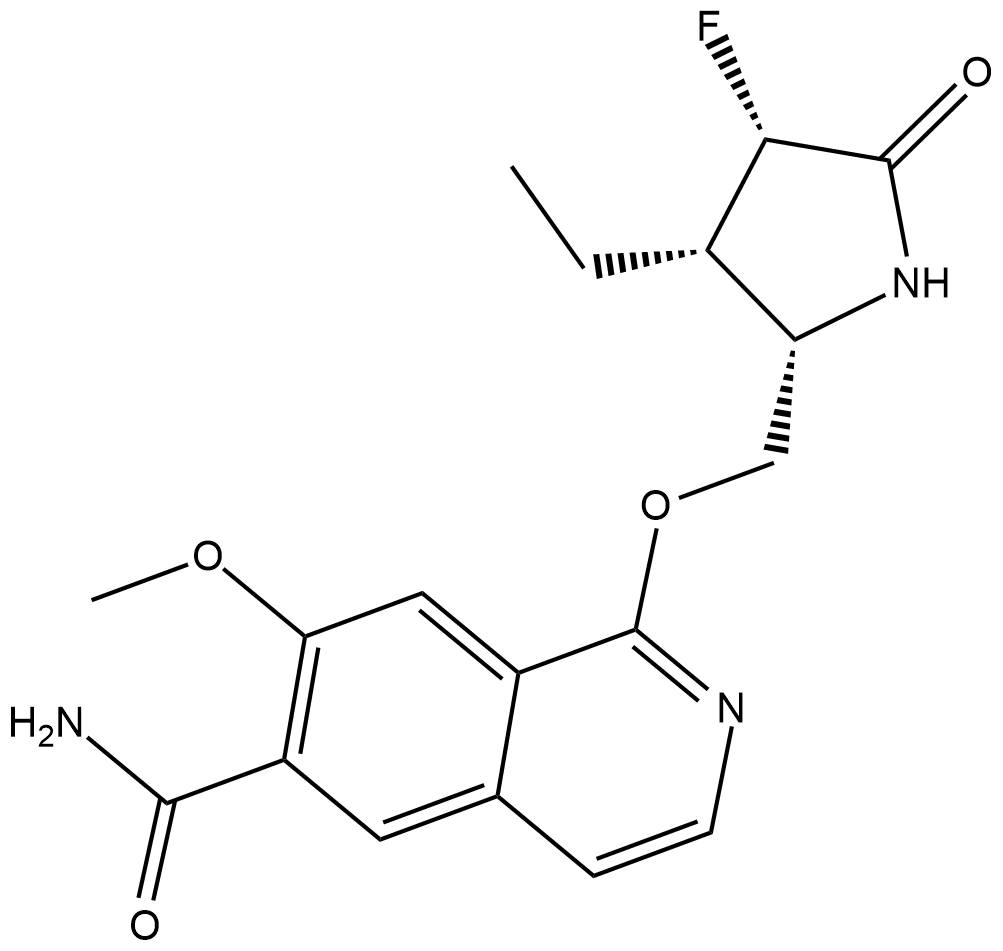
-
GC36888
PFE-360
PF-06685360
PFE-360 (PF-06685360) is a potent, selective, brain penetrated and orally active leucine-rich repeat kinase 2 (LRRK2) inhibitor with a mean IC50 of 2.3 nM in vivo.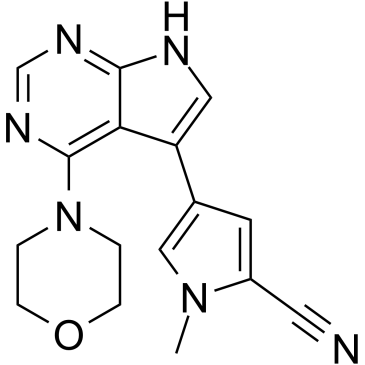
-
GC11733
PHA-665752
C-Met inhibitor,potent and ATP-competitive

-
GN10503
Piceatannol
Astringenin, transPicetannol, trans3,3',4,5'Tetrahydroxystilbene
Piceatannol (3,3′,4,5′-trans-trihydroxystilbene) is a naturally occurring hydroxylated analogue of resveratrol.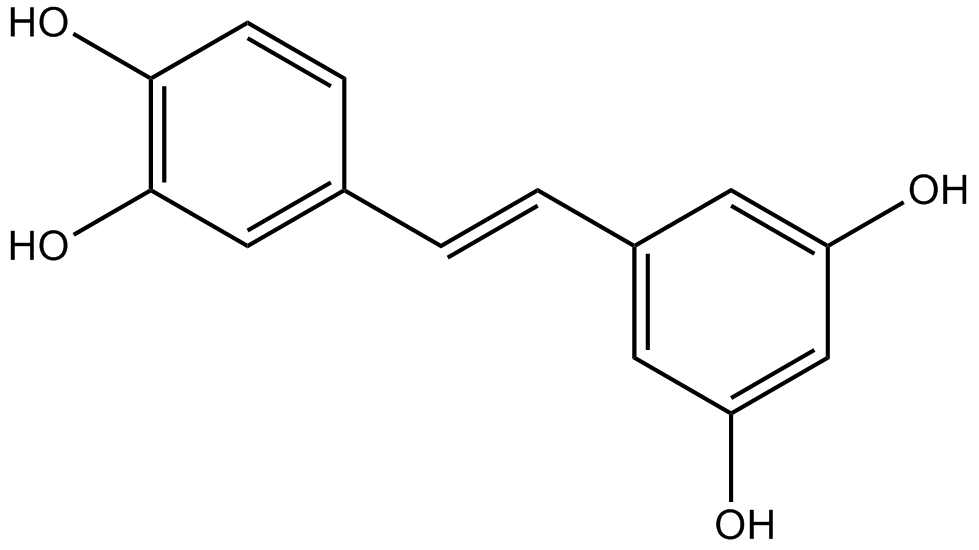
-
GC50083
PKI 166 hydrochloride
Potent EGFR-kinase inhibitor

-
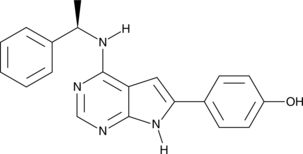
GC40915
PKI-166
An inhibitor of EGFR
-
GC17925
PKR Inhibitor
C16,GW 506033X,Protein Kinase RNA-activated
PKR Inhibitor (Compound C16) is a specific double-stranded RNA-dependent protein kinase (PKR) inhibitor.
-
GC36940
PLX5622
A CSF1R inhibitor

-
GC38836
PLX5622 hemifumarate
PLX5622 hemifumarate is a highly selective brain penetrant and orally active CSF1R inhibitor (IC50=0.016 ?M; Ki=5.9 nM).

-
GC15075
PLX647
dual inhibitor of FMS and KIT kinases

-
GC15164
PND-1186
SR 2516, VS-4718
A potent FAK inhibitor
-
GC62111
PND-1186 hydrochloride
VS-4718 hydrochloride; SR-2516 hydrochloride
PND-1186 hydrochloride (VS-4718 hydrochloride) is a potent, highly-specific and reversible inhibitor of FAK with an IC50 of 1.5 nM. PND-1186 hydrochloride selectively promotes tumor cell apoptosis.
-
GC14396
Ponatinib (AP24534)
AP 24534
Ponatinib (AP24534) (AP24534) is an orally active multi-targeted kinase inhibitor with IC50s of 0.37 nM, 1.1 nM, 1.5 nM, 2.2 nM, and 5.4 nM for Abl, PDGFRα, VEGFR2, FGFR1, and Src, respectively.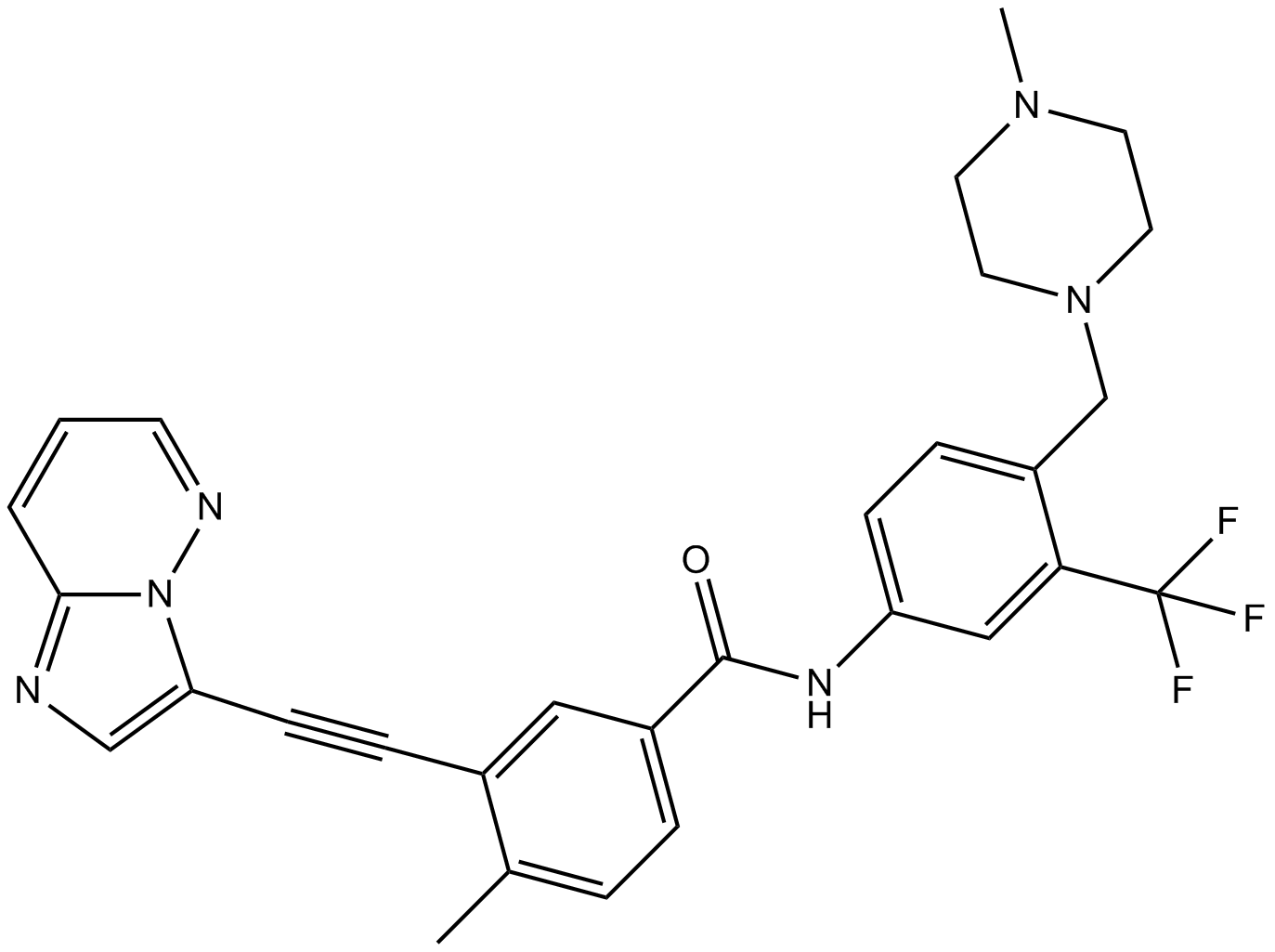
-
GC52104
Ponatinib (hydrochloride)
AP 24534
An inhibitor of native and mutant Bcr-Abl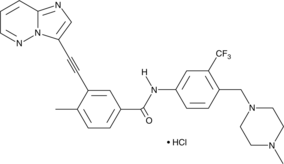
-
GC45828
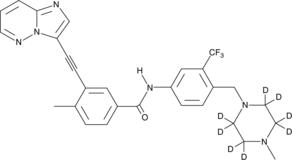
Ponatinib-d8
An internal standard for the quantification of ponatinib
-
GC69728
Ponezumab
PF-04360365; RN 1219
Ponezumab (PF-04360365) is a humanized monoclonal antibody against amyloid beta protein of the IgG2 class. Ponezumab can reduce Aβ levels in the central nervous system and improve performance in various learning and memory models in mice. Ponezumab can be used for research on Alzheimer's disease.

-
GC17916
Poziotinib
HM781-36B
A irreversible pan-HER inhibitor
-
GC17990
PP 1
AGL 1872; EI 275
Potent, selective Src family tyrosine kinase inhibitor
-
GC10344
PP 2 (AG 1879)
AGL 1879
A selective inhibitor of Src tyrosine kinases
-
GC13797
PP 3
NSC 1401
Negative control for the Src kinase inhibitor PP 2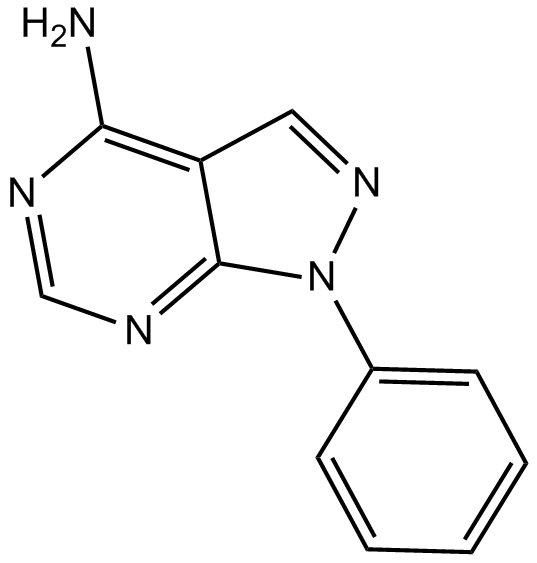
-
GC11003
PP121
Dual inhibitor of tyrosine and phosphoinositide kinases

-
GC32835
PP58
PP58 is a pyrido[2,3-d]pyrimidine-based compound that inhibits PDGFR, FGFR and Src family activities with nanomolar IC50 values.
-
GC17137
pp60 c-src (521-533) (phosphorylated)
Peptide corresponding to the pp60c-src carboxy terminal regulatory domain

-
GC12779
PPY A
Abl kinases inhibitor
-
GC16991
PQ 401
IGF1R inhibitor,potent and cell-permeable
-
GC31780
Pralsetinib (Blu667)
Pralsetinib (Blu667) (BLU-667) is a highly potent, selective RET inhibitor. Pralsetinib (Blu667) (BLU-667) inhibits WT RET, RET mutants V804L, V804M, M918T and CCDC6-RET fusion with IC50s of 0.4, 0.3, 0.4, 0.4, and 0.4 nM, respectively.
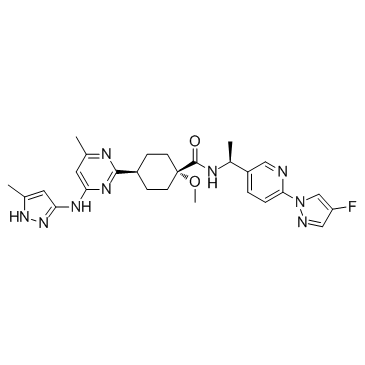
-
GC32802
PRN1371
An irreversible pan-FGFR inhibitor

-
GC30502
PRN694
PRN694 is an irreversible, highly selective and potent covalent interleukin-2-inducible T-cell kinase (ITK) and resting lymphocyte kinase (RLK) dual inhibitor with IC50s of 0.3 nM and 1.4 nM, respectively.

-
GC46208
Propentofylline
HOE 285, HWA 285
A xanthine derivative and neuroprotective agent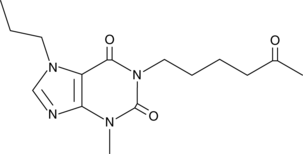
-
GC36985
PROTAC FAK degrader 1
PROTAC FAK degrader 1 is a selective and potent von Hippel-Lindau-based focal adhesion kinase (FAK) degrader with an IC50 of 6.5 nM, DC50 of 3 nM.
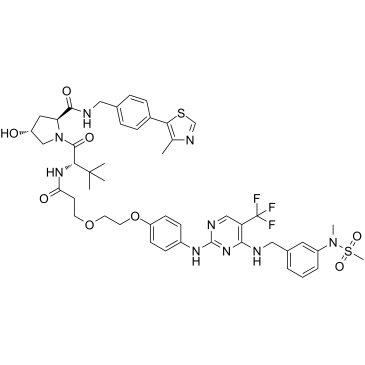
-
GC65555
PROTAC FLT-3 degrader 1
PROTAC FLT-3 degrader 1 is a von Hippel-Lindau-based PROTAC FLT-3 internal tandem duplication (ITD) degrader with an IC50 0.6 nM. Anti-proliferative activity; apoptosis induction.
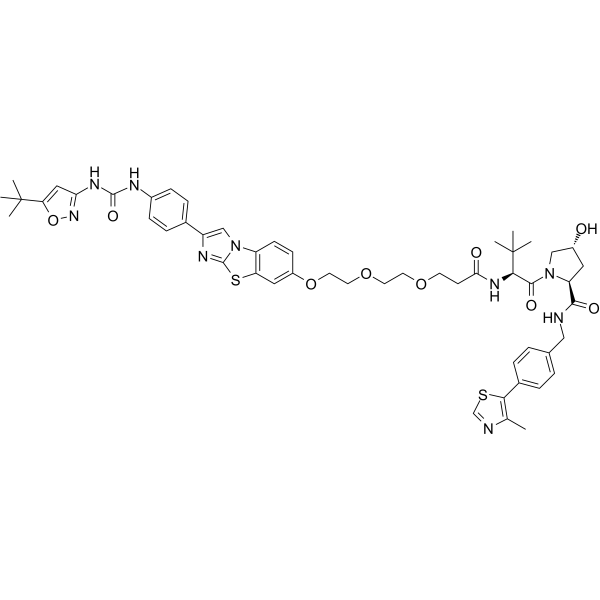
-
GC62197
PROTAC IRAK4 degrader-1
PROTAC IRAK4 degrader-1 is a Cereblon-based PROTAC interleukin-1 receptor-associated kinase 4 (IRAK4) degrader extracted from patent US20190192668A1 Compound I-210, makes 20-50%, and >50% IRAK4 degradation at 0.01, 0.1, and 1 μM in OCI-LY-10 cells, respectively.

-
GC69769
Protein kinase inhibitor 1 hydrochloride
Protein kinase inhibitor 1 hydrochloride is an effective inhibitor of HIPK2, with IC50 values of 136 and 74 nM for HIPK1 and HIPK2 respectively, and a Kd value of 9.5 nM for HIPK2.

-
GC34131
Protein kinase inhibitors 1
Protein kinase inhibitors 1 is a novel inhibitor of HIPK2 with an IC50 of 74 nM and Kd of 9.5 nM.

-
GC61217
Protein kinase inhibitors 1 hydrochloride
Protein kinase inhibitors 1 hydrochloride is a potent HIPK2 inhibitor, with IC50s of 136 and 74 nM for HIPK1 and HIPK2, and a Kd of 9.5 nM for HIPK2.
-
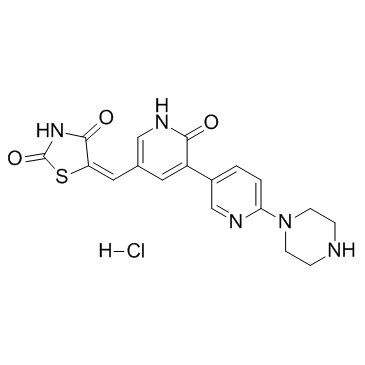
GC11321
PRT-060318
P142-76, PRT318
novel Syk inhibitor
-
GC25787
PRT-060318 2HCl
PRT318
PRT-060318 (PRT318) is a novel selective inhibitor of the Syk tyrosine kinase with an IC50 of 4 nM, as an approach to HIT treatment.
-
GC31819
PRT062607 (P505-15)
P505-15, PRT062607, PRT2607
PRT062607 (P505-15)(P505-15; PRT-2607; BIIB-057) is a highly specific and potent inhibitor of Syk with IC50 of 1-2 nM; >80-fold selective for Syk than Fgr, Lyn, FAK, Pyk2 and Zap70.
-
GC10499
PRT062607 Hydrochloride
PRT 062607 hydrochloride;PRT-062607 hydrochloride

-
GC65335
PTC299
PTC-299
PTC299 is an orally active inhibitor of VEGFA mRNA translation that selectively inhibits VEGF protein synthesis at the post-transcriptional level. PTC299 is also a potent inhibitor of dihydroorotate dehydrogenase (DHODH). PTC299 shows good oral bioavailability and lack of off-target kinase inhibition and myelosuppression. PTC299 can be useful for the research of hematologic malignancies.
-
GC32733
Pyrotinib (SHR-1258)
Pyrotinib (SHR-1258) (SHR-1258) is a potent and selective EGFR/HER2 dual inhibitor with IC50s of 13 and 38 nM, respectively.

-
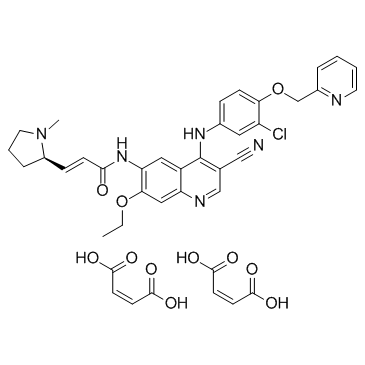
GC32989
Pyrotinib dimaleate (SHR-1258 dimaleate)
Pyrotinib dimaleate (SHR-1258 dimaleate) (SHR-1258 dimaleate) is a potent and selective EGFR/HER2 dual inhibitor with IC50s of 13 and 38 nM, respectively.
-
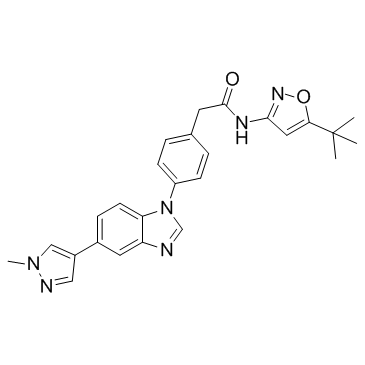
GC37047
Pz-1
Pz-1 is a potent RET and VEGFR2 inhibitor with IC50s of less than 1 nM for both wild type kinases.
-
GC17615
Quizartinib (AC220)
AC220
Quizartinib (AC220) (AC220) is an orally active, highly selective and potent second-generation type II FLT3 tyrosine kinase inhibitor, with a Kd of 1.6 nM. Quizartinib (AC220) inhibits wild-type FLT3 and FLT3-ITD autophosphorylation in MV4-11 cells with IC50s of 4.2 and 1.1 nM, respectively. Quizartinib (AC220) can be linked to the VHL ligand via an optimized linker to form a PROTAC FLT3 degrader. Quizartinib (AC220) induces apoptosis.
-
GC33874
R112
A Syk inhibitor
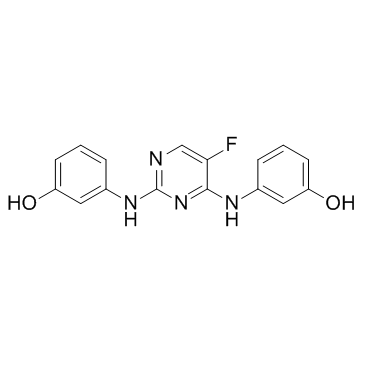
-
GC12857
R1530
A multi-kinase inhibitor

-
GC16796
R406
SYK inhibitor,potent and ATP-competitive
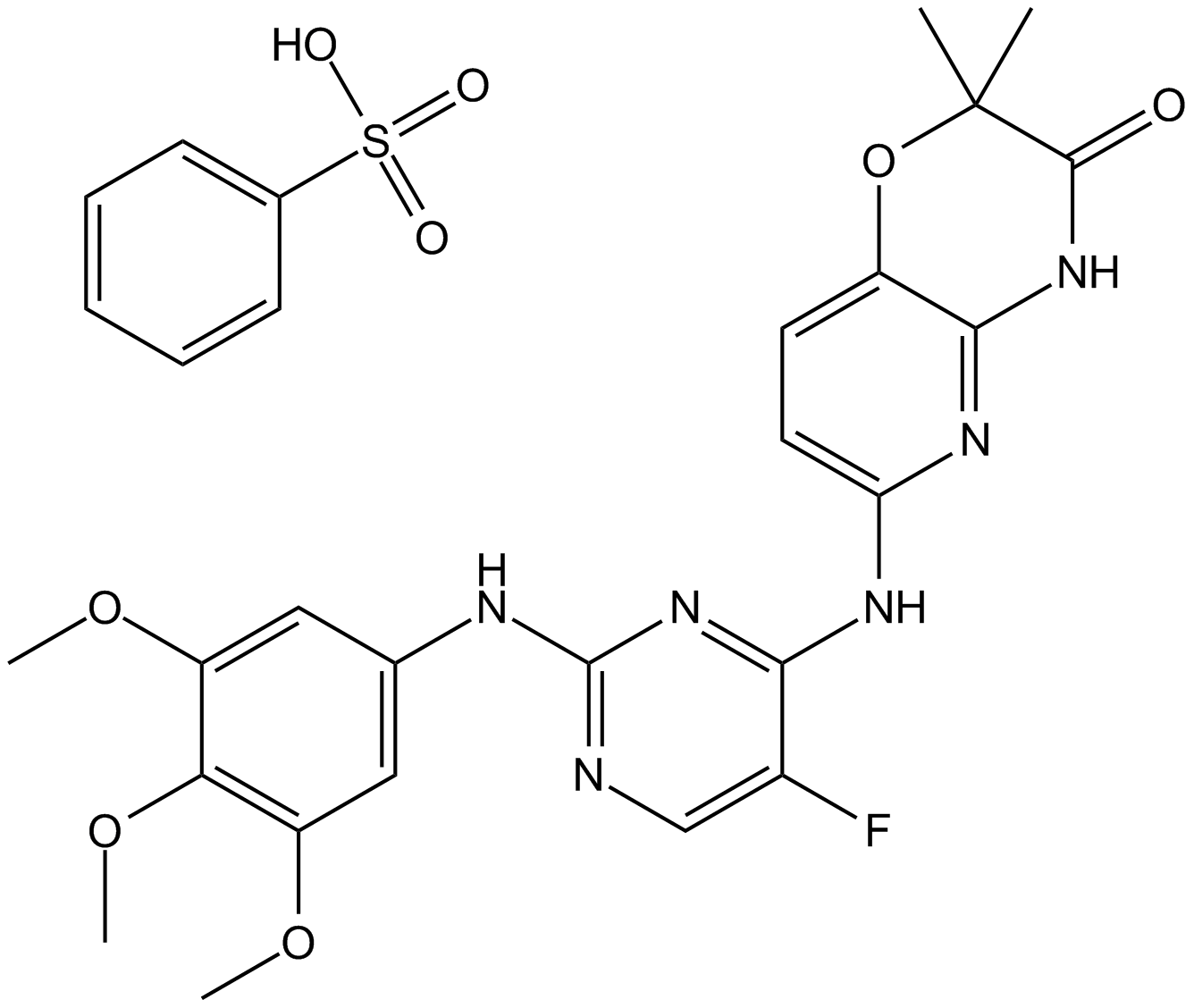
-
GC15658
R406(free base)
Syk inhibitor
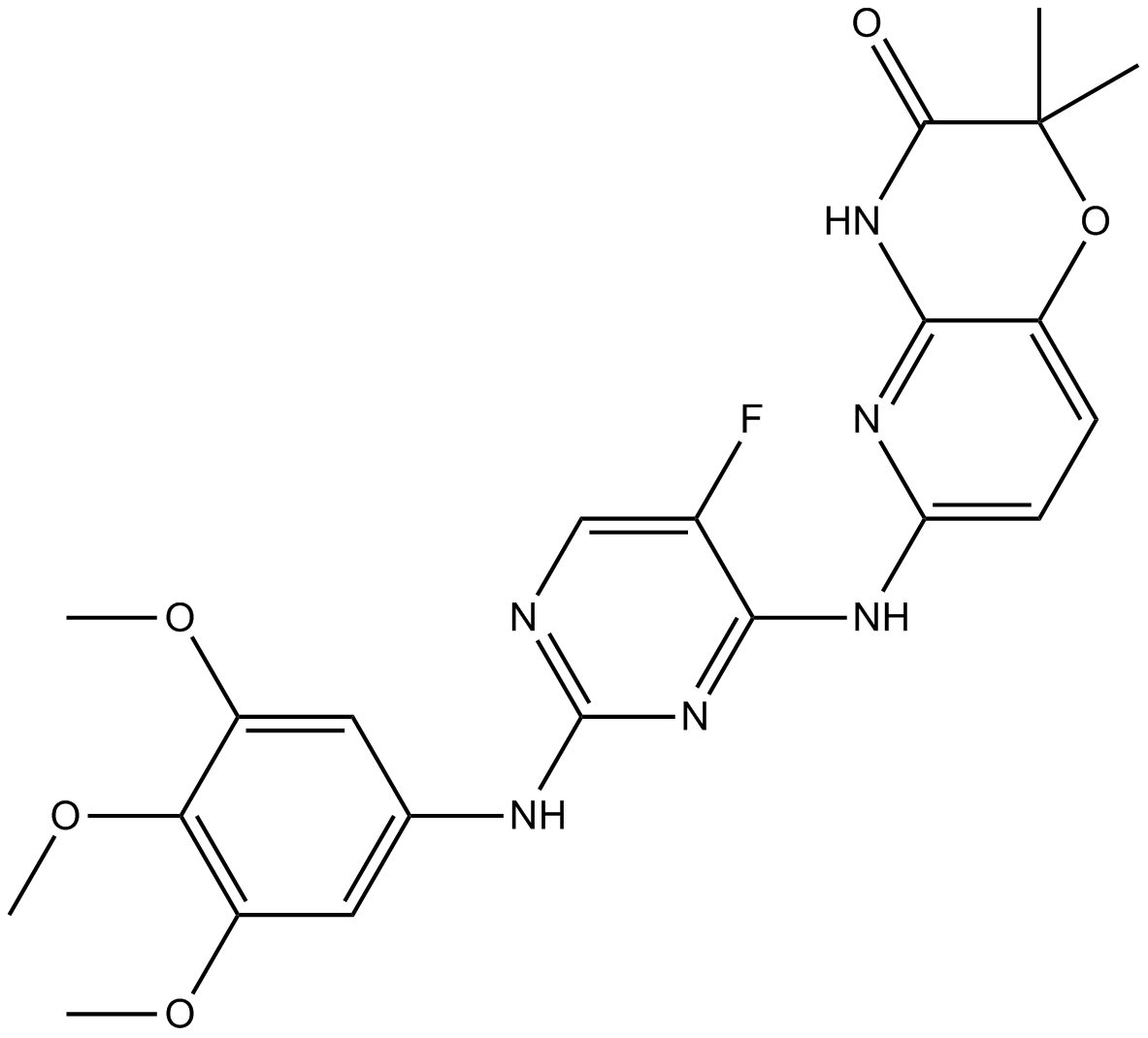
-
GC17618
R428
R-428;R 428;BGB324
R428 (R428) is a potent and selective inhibitor of Axl with an IC50 of 14 nM.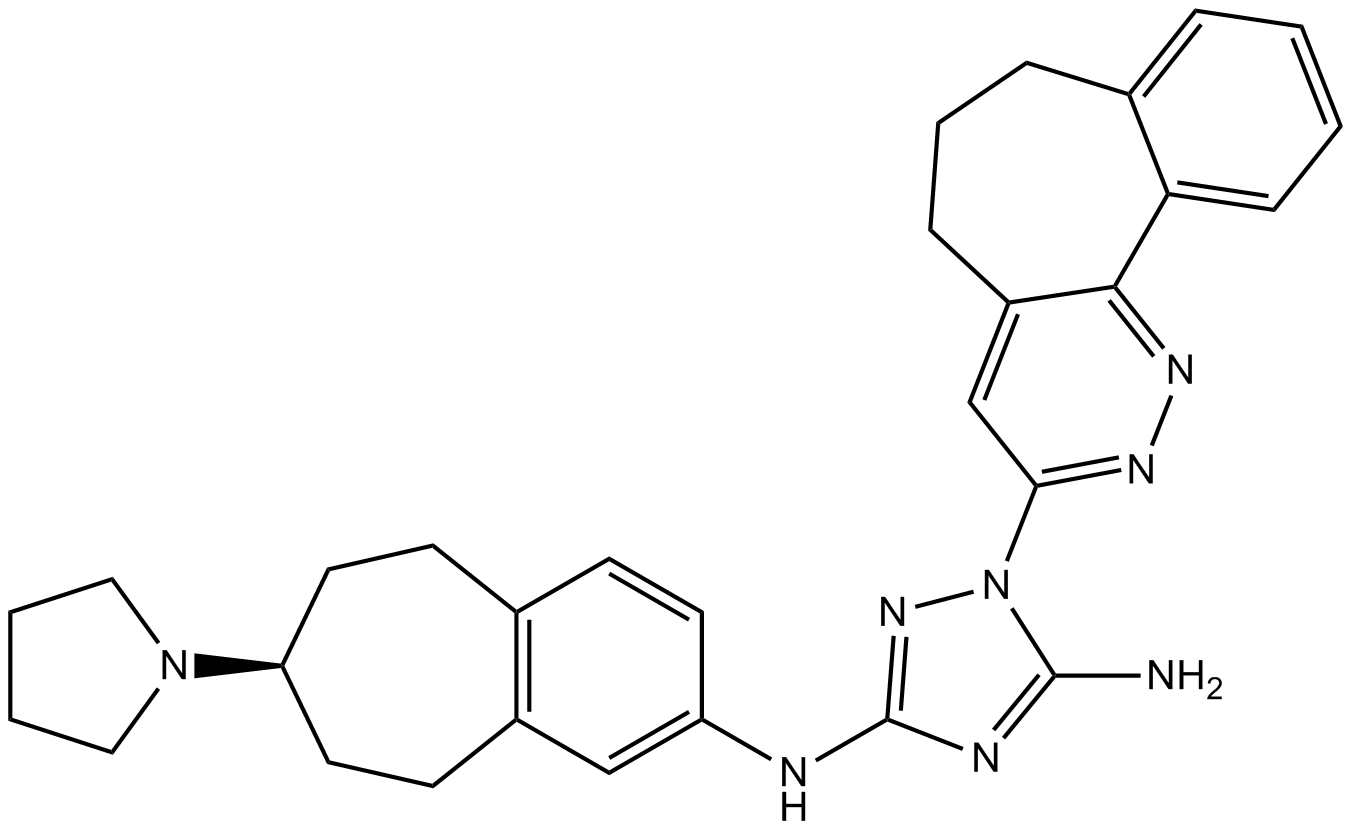
-
GC11811
R788 disodium
Fostamatinib Disodium Hexahydrate;R 788;R-788
R788 disodium (R788 Disodium) is the oral prodrug of the active compound R406.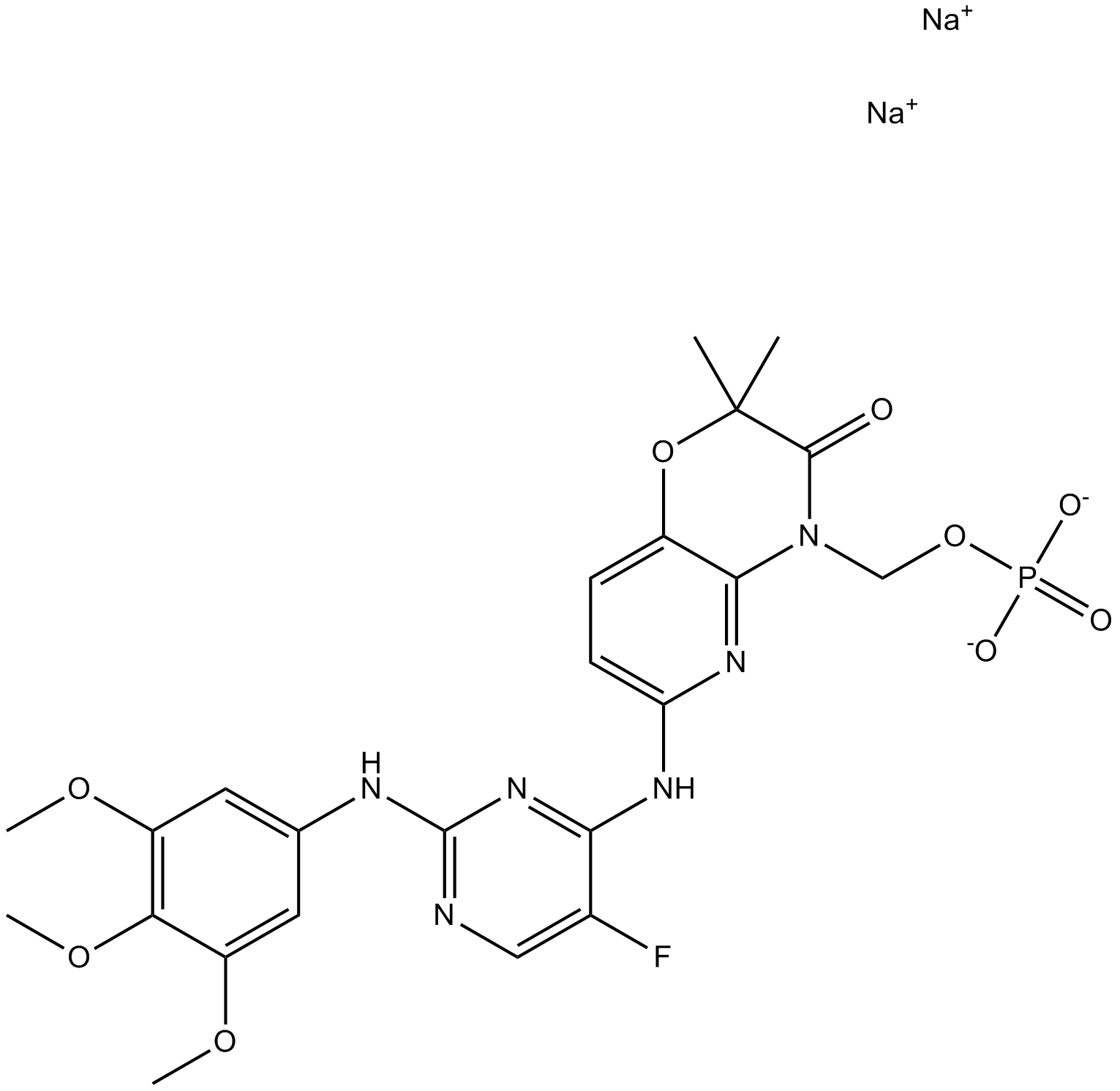
-
GC15709
R788 disodium hexahydrate
Fostamatinib (R788) disodium hexahydrate is the oral prodrug of the active compound R406.
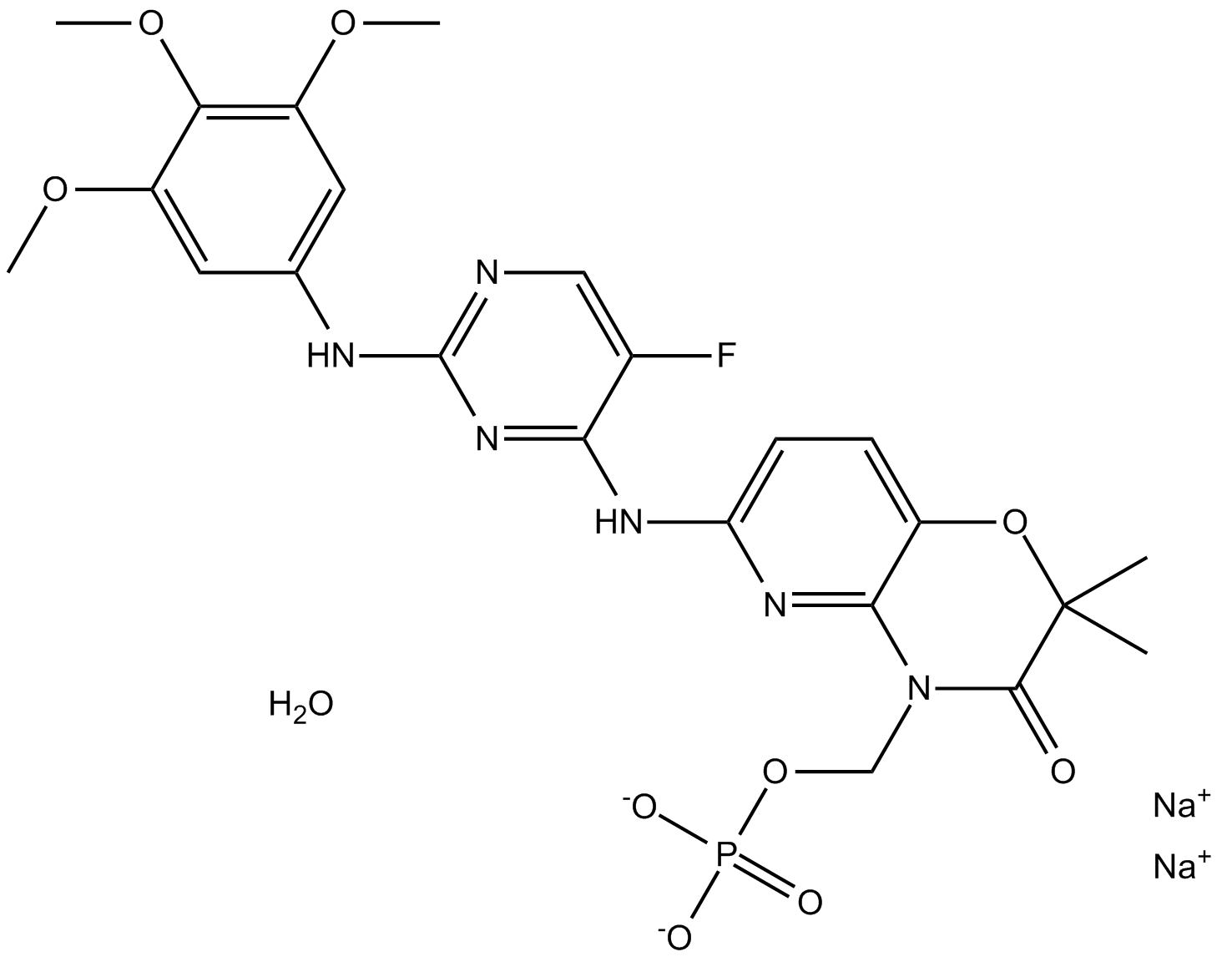
-
GC33271
R916562
R916562 is an orally active and selective Axl/VEGF-R2 inhibitor with IC50s of 136 nM and 24 nM, respectively. R916562 has anti-angiogenesis and anti-metastasis.

-
GC11140
Radotinib(IY-5511)
IY-5511
Bcr-Abl tyrosine kinase inhibitor
-
GC15818
RAF265
CHIR-265;RAF 265;RAF-265;CHIR265
Multiple intracellular kinases inhibitor
-
GC19534
Ramucirumab
LY3009806
Ramucirumab is a fully human monoclonal antibody (IgG1).

-
GC44806
Ras Inhibitory Peptide
Sos SH3 Domain Inhibitor
Son of sevenless homolog 1 (Sos1) is a guanine nucleotide exchange factor (GEF) that directs the exchange of Ras-GDP to Ras-GTP by binding to SH3 domains of the growth factor receptor-bound protein 2 (Grb2), leading to the activation of ERK.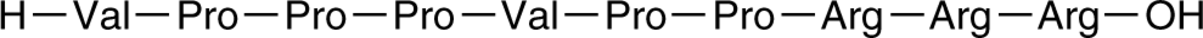
-
GC10111
Regorafenib
BAY 73-4506
A multi-kinase inhibitor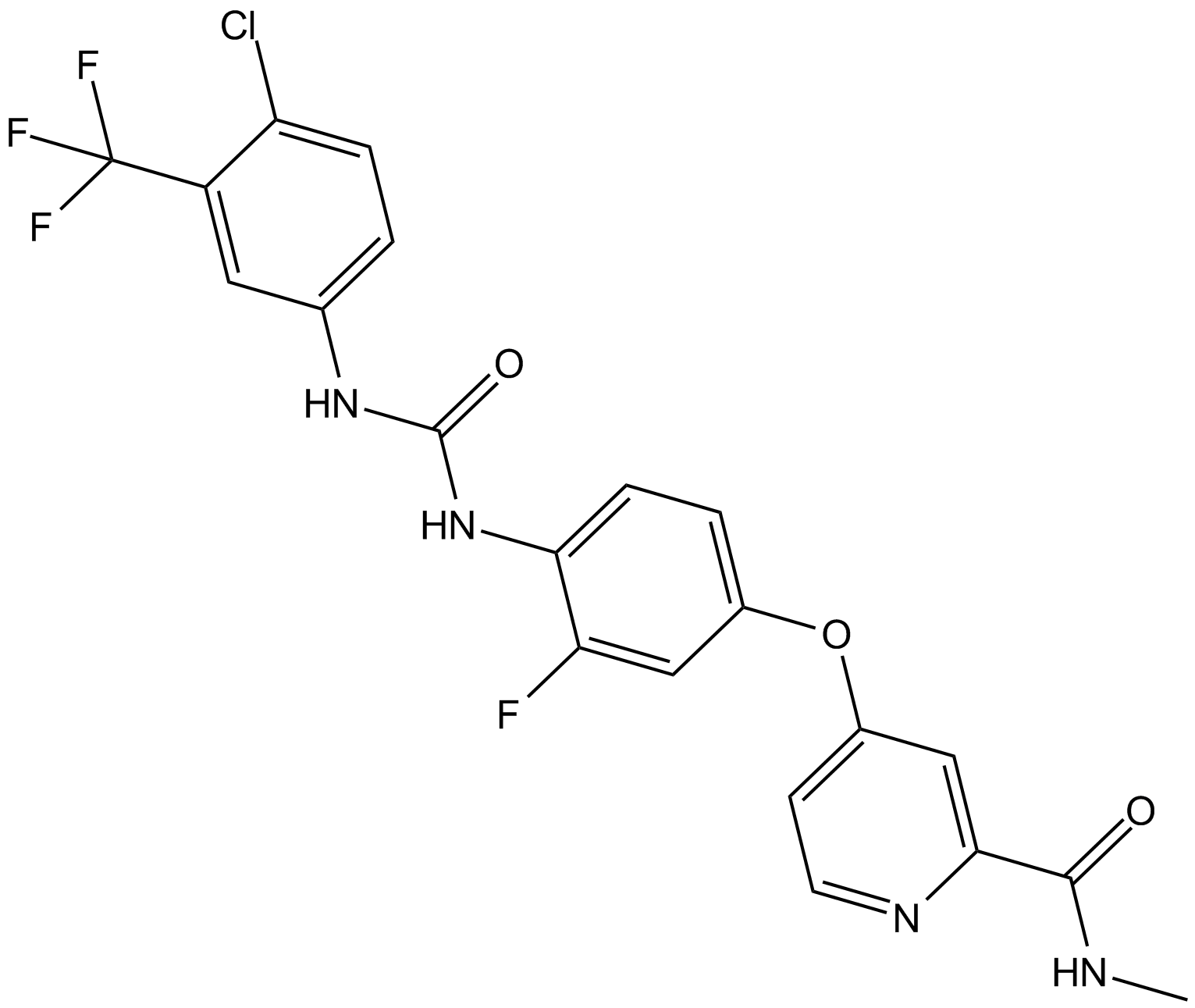
-
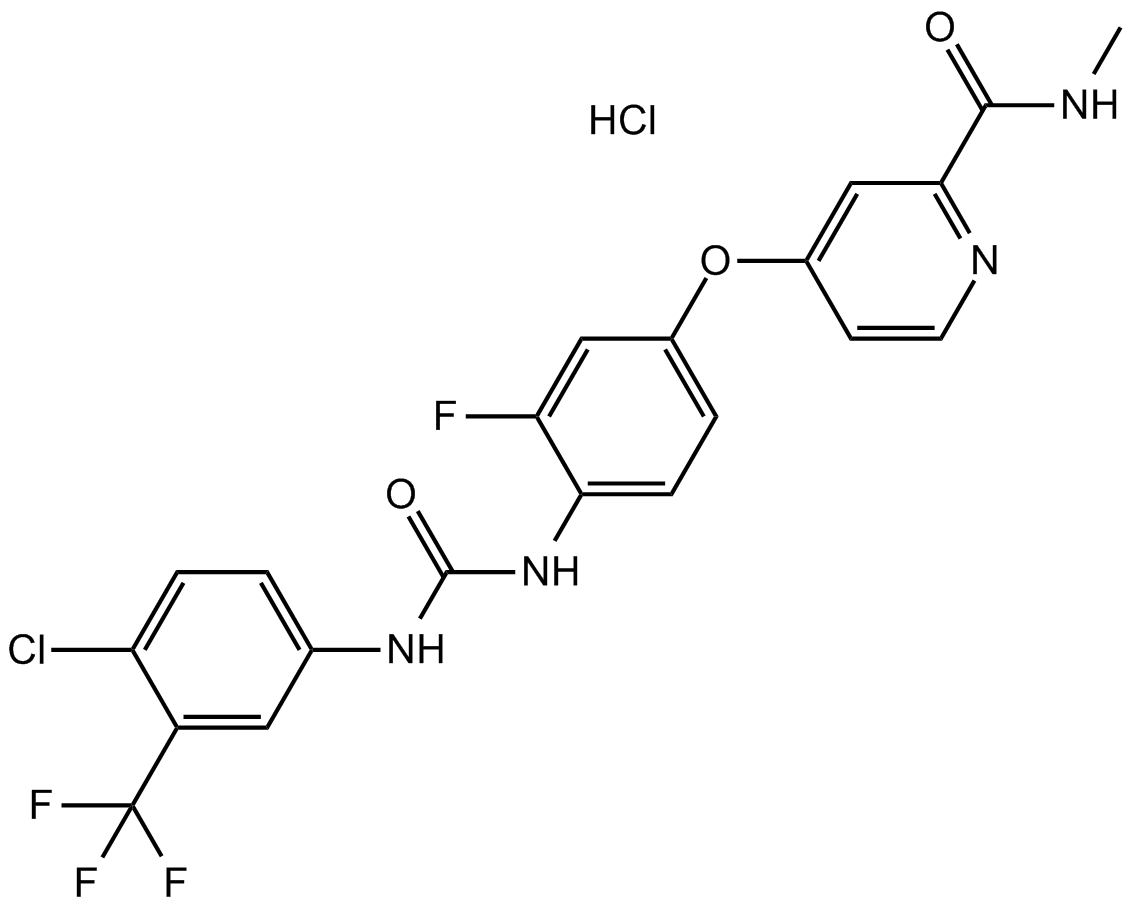
GC14606
Regorafenib hydrochloride
A multi-kinase inhibitor
-
GC14534
Regorafenib monohydrate
A multi-kinase inhibitor

-
GC40213
Regorafenib-13C-d3
Regorafenib-13C-d3 is intended for use as an internal standard for the quantification of regorafenib by GC- or LC-MS.

-
GC64895
Regorafenib-d3
BAY 73-4506-d3
Regorafenib D3 (BAY 73-4506 D3) is a deuterium labeled Regorafenib. Regorafenib is a multi-targeted receptor tyrosine kinase inhibitor.
-
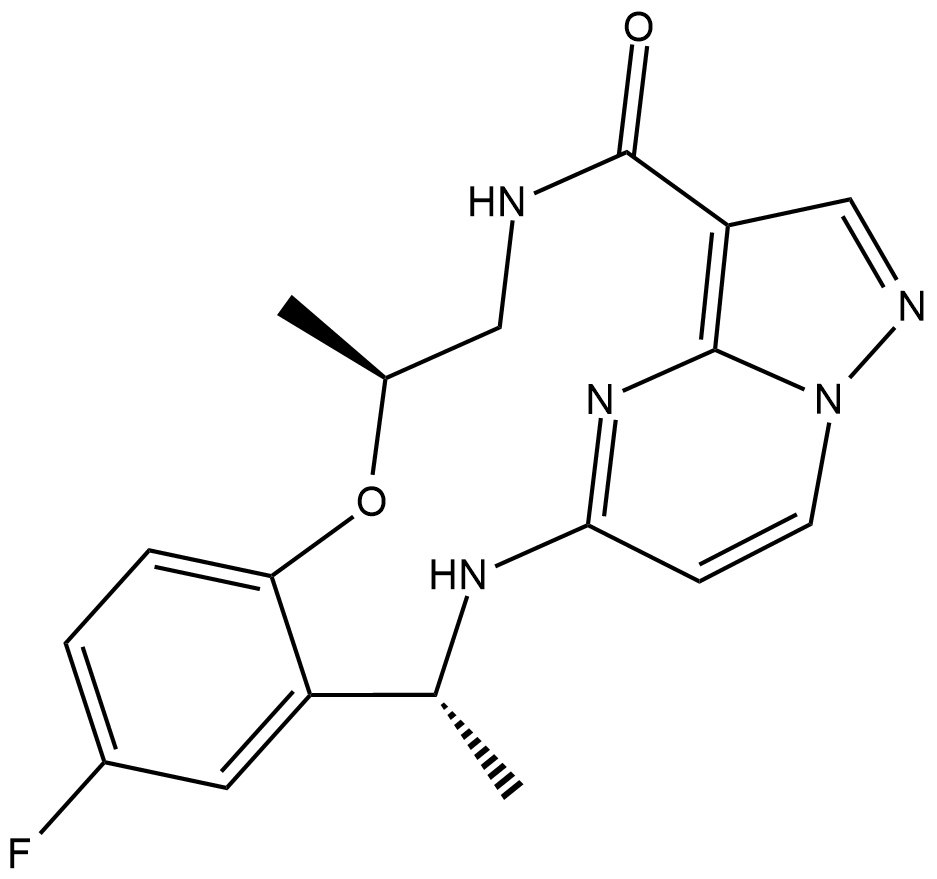
GC19362
Repotrectinib
TPX-0005 is a potent ALK/ROS1/TRK inhibitor, with IC50 of 5.3 nM, 1.01 nM, 1.26 nM and 1.08 nM for SRC, WT ALK, ALK G1202R and ALK L1196M, respectively.
-
GC67917
RET-IN-7

-
GC41467
Reveromycin D
Reveromycin D is a bacterial metabolite originally isolated from Streptomyces.

-
GC62341
Rezivertinib
BPI-7711
Rezivertinib (BPI-7711) is an orally active, highly selective and irreversible third-generation EGFR tyrosine kinase inhibitor (TKI). Rezivertinib exhibits high potency against the common activation EGFR and the resistance T790M mutations. Rezivertinib has excellent central nervous system (CNS) penetration and has antitumor activity.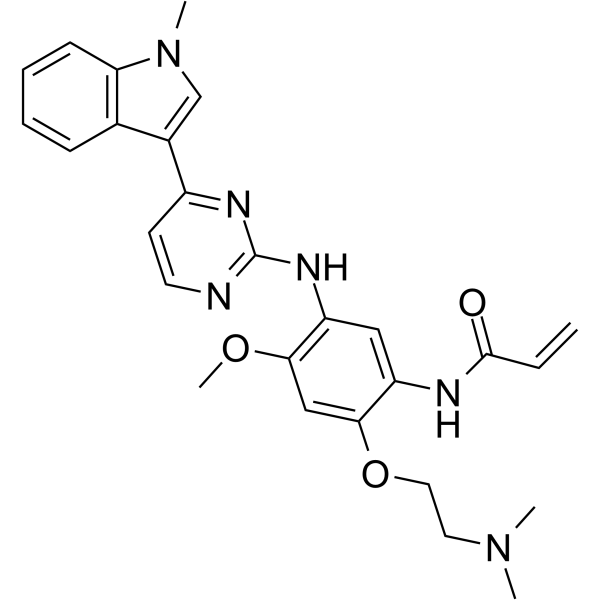
-
GC12038
RG 13022
Tyrphostin RG13022
EGFR tyrosine kinase inhibitor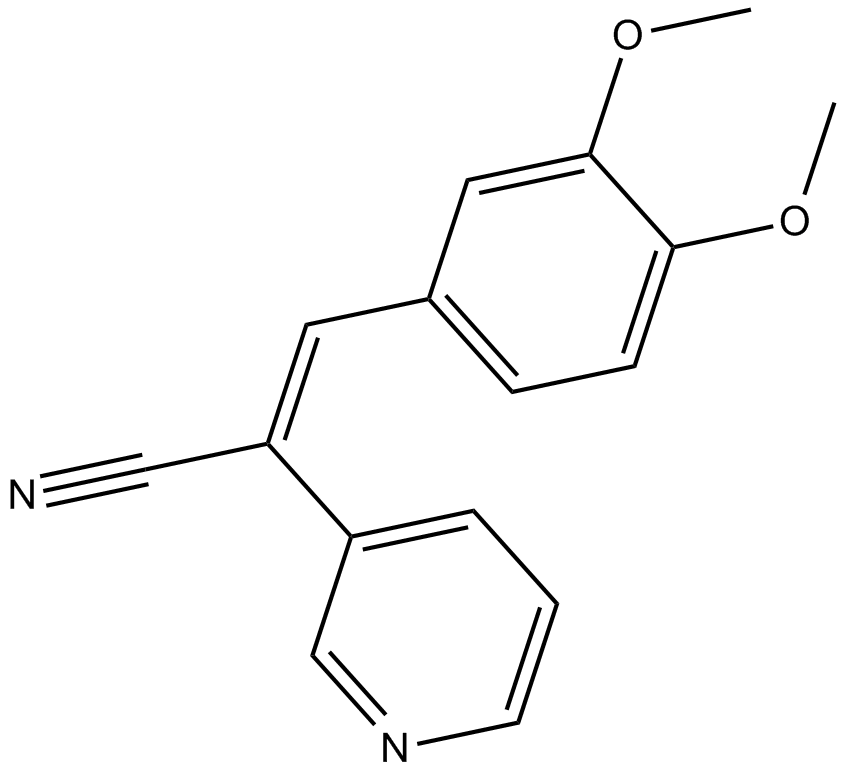
-
GC10217
RG-14620
Tyrphostin RG14620
inhibitor of epidermal growth factor (EGF) receptor kinase
-
GN10784
Rhoifolin
Apigenin 7-O-Neohesperidoside

-
GC69817
Rilotumumab
AMG 102
Rilotumumab (AMG 102) is a monoclonal antibody that targets the hepatocyte growth factor (HGF), inhibiting HGF/MET-driven signaling. Rilotumumab has anti-tumor activity and is being studied for use in castration-resistant prostate cancer (CRPC) and solid tumors.
-
GC37538
Ripretinib
DCC-2618
Ripretinib (DCC-2618) is an orally bioavailable, selective KIT and PDGFRA switch-control inhibitor. Ripretinib (DCC-2618) targets and binds to both wild-type and mutant forms of KIT and PDGFRA specifically at their switch pocket binding sites, thereby preventing the switch from inactive to active conformations of these kinases and inactivating their wild-type and mutant forms. Ripretinib (DCC-2618) also inhibits multiple other kinase targets, such as FLT3 and KDR (or VEGFR-2). DCC-2618 exerts antineoplastic effect and induces apoptosis.
-
GC40547
RK-20448
RK-20448 is an ATP-competitive inhibitor of Lck, Src, KDR/VEGF2R, and Tie-2 (IC50s = 0.24, 1.19, 10.74, and 5.85 μM, respectively).



